
Butyrivibrio crossotus CAG:259
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Butyrivibrio; environmental samples
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
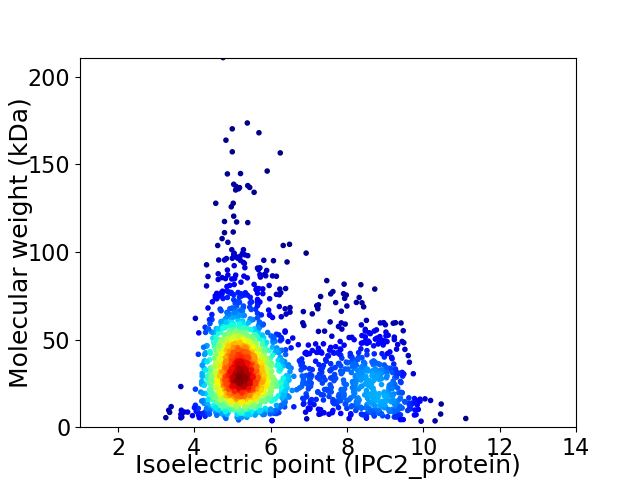
Virtual 2D-PAGE plot for 1976 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5LWY2|R5LWY2_9FIRM YitL protein OS=Butyrivibrio crossotus CAG:259 OX=1263062 GN=BN569_00806 PE=3 SV=1
MM1 pKa = 7.76KK2 pKa = 10.11EE3 pKa = 3.82PEE5 pKa = 3.74IDD7 pKa = 3.48IEE9 pKa = 4.18EE10 pKa = 4.74DD11 pKa = 3.34STDD14 pKa = 3.36IEE16 pKa = 4.53VIEE19 pKa = 4.83IDD21 pKa = 3.91SDD23 pKa = 4.27TIDD26 pKa = 3.62GAAADD31 pKa = 4.42SDD33 pKa = 4.16VASNIQNVSEE43 pKa = 4.34EE44 pKa = 4.2NDD46 pKa = 3.68PPVQPDD52 pKa = 3.51NEE54 pKa = 4.27IEE56 pKa = 4.23NTDD59 pKa = 3.34TTEE62 pKa = 3.86QTTSDD67 pKa = 4.39DD68 pKa = 3.71DD69 pKa = 4.38FEE71 pKa = 6.31IIPAEE76 pKa = 4.25AVPDD80 pKa = 4.73DD81 pKa = 4.12YY82 pKa = 11.88DD83 pKa = 4.51NYY85 pKa = 10.66ILTPDD90 pKa = 3.13NRR92 pKa = 11.84KK93 pKa = 8.66FHH95 pKa = 5.81ITKK98 pKa = 9.97FDD100 pKa = 2.95IRR102 pKa = 11.84RR103 pKa = 11.84YY104 pKa = 9.64IIIFILTCVMVFCMYY119 pKa = 10.38QIISRR124 pKa = 11.84YY125 pKa = 8.76IGYY128 pKa = 10.09ARR130 pKa = 11.84EE131 pKa = 3.8RR132 pKa = 11.84RR133 pKa = 11.84HH134 pKa = 5.89NSEE137 pKa = 3.85INNSVIEE144 pKa = 4.59EE145 pKa = 4.19NTSTPDD151 pKa = 4.25LSTDD155 pKa = 3.2TYY157 pKa = 11.77KK158 pKa = 11.13NINPGLAIDD167 pKa = 4.48FDD169 pKa = 4.45KK170 pKa = 11.16LLQINEE176 pKa = 4.24ACAGWIQIPSVGISYY191 pKa = 8.87PIVQGPDD198 pKa = 2.56NDD200 pKa = 4.23YY201 pKa = 10.75YY202 pKa = 11.43LHH204 pKa = 6.9YY205 pKa = 10.64DD206 pKa = 3.49ISQNEE211 pKa = 3.99AWSGAIYY218 pKa = 10.54LDD220 pKa = 3.65YY221 pKa = 11.04RR222 pKa = 11.84SDD224 pKa = 3.46RR225 pKa = 11.84SLDD228 pKa = 3.58EE229 pKa = 3.76QHH231 pKa = 5.27FTIYY235 pKa = 9.58GHH237 pKa = 6.48HH238 pKa = 6.56MNDD241 pKa = 2.74GSMFANLLKK250 pKa = 10.66YY251 pKa = 10.38DD252 pKa = 4.39DD253 pKa = 4.35PEE255 pKa = 4.65FYY257 pKa = 10.54QKK259 pKa = 10.79NQDD262 pKa = 3.14NFNNYY267 pKa = 8.74IYY269 pKa = 9.74IYY271 pKa = 9.8QKK273 pKa = 10.89GRR275 pKa = 11.84VLVYY279 pKa = 9.98QIFSVVDD286 pKa = 3.39TYY288 pKa = 11.83YY289 pKa = 11.25DD290 pKa = 3.5SDD292 pKa = 4.11PEE294 pKa = 4.26SFLVLISDD302 pKa = 4.55DD303 pKa = 3.97EE304 pKa = 4.67TKK306 pKa = 10.63KK307 pKa = 10.95SYY309 pKa = 10.95LNHH312 pKa = 6.22IRR314 pKa = 11.84QKK316 pKa = 10.32QLYY319 pKa = 5.64EE320 pKa = 3.69TGYY323 pKa = 10.6FADD326 pKa = 3.82TNDD329 pKa = 4.26RR330 pKa = 11.84LLTLYY335 pKa = 9.21TCQSDD340 pKa = 3.86STSKK344 pKa = 9.78EE345 pKa = 3.33RR346 pKa = 11.84HH347 pKa = 4.69MVHH350 pKa = 6.77AKK352 pKa = 10.41LIAEE356 pKa = 4.63ISNN359 pKa = 3.56
MM1 pKa = 7.76KK2 pKa = 10.11EE3 pKa = 3.82PEE5 pKa = 3.74IDD7 pKa = 3.48IEE9 pKa = 4.18EE10 pKa = 4.74DD11 pKa = 3.34STDD14 pKa = 3.36IEE16 pKa = 4.53VIEE19 pKa = 4.83IDD21 pKa = 3.91SDD23 pKa = 4.27TIDD26 pKa = 3.62GAAADD31 pKa = 4.42SDD33 pKa = 4.16VASNIQNVSEE43 pKa = 4.34EE44 pKa = 4.2NDD46 pKa = 3.68PPVQPDD52 pKa = 3.51NEE54 pKa = 4.27IEE56 pKa = 4.23NTDD59 pKa = 3.34TTEE62 pKa = 3.86QTTSDD67 pKa = 4.39DD68 pKa = 3.71DD69 pKa = 4.38FEE71 pKa = 6.31IIPAEE76 pKa = 4.25AVPDD80 pKa = 4.73DD81 pKa = 4.12YY82 pKa = 11.88DD83 pKa = 4.51NYY85 pKa = 10.66ILTPDD90 pKa = 3.13NRR92 pKa = 11.84KK93 pKa = 8.66FHH95 pKa = 5.81ITKK98 pKa = 9.97FDD100 pKa = 2.95IRR102 pKa = 11.84RR103 pKa = 11.84YY104 pKa = 9.64IIIFILTCVMVFCMYY119 pKa = 10.38QIISRR124 pKa = 11.84YY125 pKa = 8.76IGYY128 pKa = 10.09ARR130 pKa = 11.84EE131 pKa = 3.8RR132 pKa = 11.84RR133 pKa = 11.84HH134 pKa = 5.89NSEE137 pKa = 3.85INNSVIEE144 pKa = 4.59EE145 pKa = 4.19NTSTPDD151 pKa = 4.25LSTDD155 pKa = 3.2TYY157 pKa = 11.77KK158 pKa = 11.13NINPGLAIDD167 pKa = 4.48FDD169 pKa = 4.45KK170 pKa = 11.16LLQINEE176 pKa = 4.24ACAGWIQIPSVGISYY191 pKa = 8.87PIVQGPDD198 pKa = 2.56NDD200 pKa = 4.23YY201 pKa = 10.75YY202 pKa = 11.43LHH204 pKa = 6.9YY205 pKa = 10.64DD206 pKa = 3.49ISQNEE211 pKa = 3.99AWSGAIYY218 pKa = 10.54LDD220 pKa = 3.65YY221 pKa = 11.04RR222 pKa = 11.84SDD224 pKa = 3.46RR225 pKa = 11.84SLDD228 pKa = 3.58EE229 pKa = 3.76QHH231 pKa = 5.27FTIYY235 pKa = 9.58GHH237 pKa = 6.48HH238 pKa = 6.56MNDD241 pKa = 2.74GSMFANLLKK250 pKa = 10.66YY251 pKa = 10.38DD252 pKa = 4.39DD253 pKa = 4.35PEE255 pKa = 4.65FYY257 pKa = 10.54QKK259 pKa = 10.79NQDD262 pKa = 3.14NFNNYY267 pKa = 8.74IYY269 pKa = 9.74IYY271 pKa = 9.8QKK273 pKa = 10.89GRR275 pKa = 11.84VLVYY279 pKa = 9.98QIFSVVDD286 pKa = 3.39TYY288 pKa = 11.83YY289 pKa = 11.25DD290 pKa = 3.5SDD292 pKa = 4.11PEE294 pKa = 4.26SFLVLISDD302 pKa = 4.55DD303 pKa = 3.97EE304 pKa = 4.67TKK306 pKa = 10.63KK307 pKa = 10.95SYY309 pKa = 10.95LNHH312 pKa = 6.22IRR314 pKa = 11.84QKK316 pKa = 10.32QLYY319 pKa = 5.64EE320 pKa = 3.69TGYY323 pKa = 10.6FADD326 pKa = 3.82TNDD329 pKa = 4.26RR330 pKa = 11.84LLTLYY335 pKa = 9.21TCQSDD340 pKa = 3.86STSKK344 pKa = 9.78EE345 pKa = 3.33RR346 pKa = 11.84HH347 pKa = 4.69MVHH350 pKa = 6.77AKK352 pKa = 10.41LIAEE356 pKa = 4.63ISNN359 pKa = 3.56
Molecular weight: 41.62 kDa
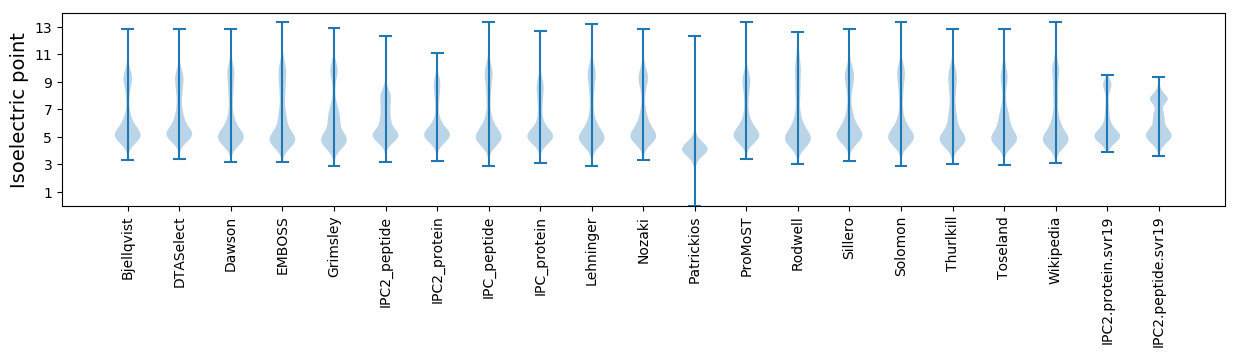
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5LUI2|R5LUI2_9FIRM Oxidoreductase Gfo/Idh/MocA family OS=Butyrivibrio crossotus CAG:259 OX=1263062 GN=BN569_00355 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.27VHH16 pKa = 6.08GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.27VHH16 pKa = 6.08GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 4.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
629432 |
30 |
1985 |
318.5 |
35.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.744 ± 0.058 | 1.41 ± 0.022 |
6.326 ± 0.045 | 7.229 ± 0.055 |
4.154 ± 0.044 | 6.837 ± 0.056 |
1.479 ± 0.023 | 8.739 ± 0.055 |
7.849 ± 0.047 | 8.072 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.003 ± 0.025 | 5.419 ± 0.04 |
2.871 ± 0.028 | 2.165 ± 0.024 |
3.925 ± 0.035 | 6.011 ± 0.044 |
5.533 ± 0.048 | 7.084 ± 0.045 |
0.644 ± 0.018 | 4.501 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
