
Sewage associated gemycircularvirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus sewopo1; Sewage derived gemykibivirus 1
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
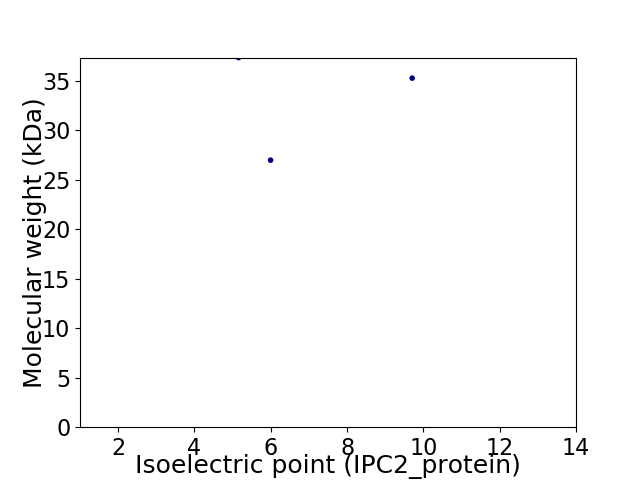
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161IVR5|A0A161IVR5_9VIRU RepA OS=Sewage associated gemycircularvirus 3 OX=1843761 PE=3 SV=1
MM1 pKa = 7.45TFRR4 pKa = 11.84FAAKK8 pKa = 10.31YY9 pKa = 10.64GLLTYY14 pKa = 10.56AQIGDD19 pKa = 3.65RR20 pKa = 11.84DD21 pKa = 3.79VEE23 pKa = 4.24EE24 pKa = 4.58FGWRR28 pKa = 11.84VSDD31 pKa = 3.48MLGSLGAEE39 pKa = 3.96CIVGRR44 pKa = 11.84EE45 pKa = 3.97SHH47 pKa = 6.97ADD49 pKa = 3.29GGLHH53 pKa = 5.1IHH55 pKa = 6.74AFFMFEE61 pKa = 4.81RR62 pKa = 11.84KK63 pKa = 8.84FQSRR67 pKa = 11.84NVRR70 pKa = 11.84VFDD73 pKa = 4.43MDD75 pKa = 3.54GCHH78 pKa = 6.22PNIVRR83 pKa = 11.84GYY85 pKa = 7.79STPEE89 pKa = 3.72DD90 pKa = 3.56GARR93 pKa = 11.84YY94 pKa = 9.33AIKK97 pKa = 10.44EE98 pKa = 3.95GDD100 pKa = 3.98VIAGGLDD107 pKa = 3.41VDD109 pKa = 4.37SLGSSVAGSKK119 pKa = 7.63TVWAQIILAEE129 pKa = 4.26SRR131 pKa = 11.84DD132 pKa = 3.92DD133 pKa = 4.9FFAACAEE140 pKa = 4.11LAPRR144 pKa = 11.84ALLCSFTSLRR154 pKa = 11.84CYY156 pKa = 10.98ADD158 pKa = 2.7WKK160 pKa = 10.01YY161 pKa = 11.04RR162 pKa = 11.84EE163 pKa = 4.58DD164 pKa = 3.75PAPYY168 pKa = 9.65RR169 pKa = 11.84HH170 pKa = 6.68PEE172 pKa = 4.08DD173 pKa = 3.74VSFDD177 pKa = 3.39TSRR180 pKa = 11.84FPEE183 pKa = 4.14LDD185 pKa = 2.56RR186 pKa = 11.84WVQDD190 pKa = 3.46SLRR193 pKa = 11.84GAQRR197 pKa = 11.84GRR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84SLILYY206 pKa = 9.32GPTKK210 pKa = 10.4LGKK213 pKa = 7.53TLWARR218 pKa = 11.84SLGNHH223 pKa = 7.12AYY225 pKa = 10.38FGGLFSMDD233 pKa = 3.49EE234 pKa = 4.55SIDD237 pKa = 3.78DD238 pKa = 2.96VDD240 pKa = 3.91YY241 pKa = 11.58AVFDD245 pKa = 4.3DD246 pKa = 4.09MQGGLKK252 pKa = 10.09FFHH255 pKa = 7.11SYY257 pKa = 10.7KK258 pKa = 10.25FWLGAQSQFYY268 pKa = 8.76VTEE271 pKa = 4.22KK272 pKa = 10.81YY273 pKa = 9.41KK274 pKa = 10.96GKK276 pKa = 10.51RR277 pKa = 11.84LVHH280 pKa = 6.05WGKK283 pKa = 9.71PCIYY287 pKa = 9.84LYY289 pKa = 10.87NHH291 pKa = 6.64NPLCDD296 pKa = 3.41EE297 pKa = 4.74GADD300 pKa = 4.1HH301 pKa = 7.86DD302 pKa = 4.56WLLGNCDD309 pKa = 3.68IVGLDD314 pKa = 4.13ADD316 pKa = 4.81DD317 pKa = 4.32SLLVPEE323 pKa = 4.95EE324 pKa = 4.32GSLGGEE330 pKa = 4.03RR331 pKa = 4.37
MM1 pKa = 7.45TFRR4 pKa = 11.84FAAKK8 pKa = 10.31YY9 pKa = 10.64GLLTYY14 pKa = 10.56AQIGDD19 pKa = 3.65RR20 pKa = 11.84DD21 pKa = 3.79VEE23 pKa = 4.24EE24 pKa = 4.58FGWRR28 pKa = 11.84VSDD31 pKa = 3.48MLGSLGAEE39 pKa = 3.96CIVGRR44 pKa = 11.84EE45 pKa = 3.97SHH47 pKa = 6.97ADD49 pKa = 3.29GGLHH53 pKa = 5.1IHH55 pKa = 6.74AFFMFEE61 pKa = 4.81RR62 pKa = 11.84KK63 pKa = 8.84FQSRR67 pKa = 11.84NVRR70 pKa = 11.84VFDD73 pKa = 4.43MDD75 pKa = 3.54GCHH78 pKa = 6.22PNIVRR83 pKa = 11.84GYY85 pKa = 7.79STPEE89 pKa = 3.72DD90 pKa = 3.56GARR93 pKa = 11.84YY94 pKa = 9.33AIKK97 pKa = 10.44EE98 pKa = 3.95GDD100 pKa = 3.98VIAGGLDD107 pKa = 3.41VDD109 pKa = 4.37SLGSSVAGSKK119 pKa = 7.63TVWAQIILAEE129 pKa = 4.26SRR131 pKa = 11.84DD132 pKa = 3.92DD133 pKa = 4.9FFAACAEE140 pKa = 4.11LAPRR144 pKa = 11.84ALLCSFTSLRR154 pKa = 11.84CYY156 pKa = 10.98ADD158 pKa = 2.7WKK160 pKa = 10.01YY161 pKa = 11.04RR162 pKa = 11.84EE163 pKa = 4.58DD164 pKa = 3.75PAPYY168 pKa = 9.65RR169 pKa = 11.84HH170 pKa = 6.68PEE172 pKa = 4.08DD173 pKa = 3.74VSFDD177 pKa = 3.39TSRR180 pKa = 11.84FPEE183 pKa = 4.14LDD185 pKa = 2.56RR186 pKa = 11.84WVQDD190 pKa = 3.46SLRR193 pKa = 11.84GAQRR197 pKa = 11.84GRR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84SLILYY206 pKa = 9.32GPTKK210 pKa = 10.4LGKK213 pKa = 7.53TLWARR218 pKa = 11.84SLGNHH223 pKa = 7.12AYY225 pKa = 10.38FGGLFSMDD233 pKa = 3.49EE234 pKa = 4.55SIDD237 pKa = 3.78DD238 pKa = 2.96VDD240 pKa = 3.91YY241 pKa = 11.58AVFDD245 pKa = 4.3DD246 pKa = 4.09MQGGLKK252 pKa = 10.09FFHH255 pKa = 7.11SYY257 pKa = 10.7KK258 pKa = 10.25FWLGAQSQFYY268 pKa = 8.76VTEE271 pKa = 4.22KK272 pKa = 10.81YY273 pKa = 9.41KK274 pKa = 10.96GKK276 pKa = 10.51RR277 pKa = 11.84LVHH280 pKa = 6.05WGKK283 pKa = 9.71PCIYY287 pKa = 9.84LYY289 pKa = 10.87NHH291 pKa = 6.64NPLCDD296 pKa = 3.41EE297 pKa = 4.74GADD300 pKa = 4.1HH301 pKa = 7.86DD302 pKa = 4.56WLLGNCDD309 pKa = 3.68IVGLDD314 pKa = 4.13ADD316 pKa = 4.81DD317 pKa = 4.32SLLVPEE323 pKa = 4.95EE324 pKa = 4.32GSLGGEE330 pKa = 4.03RR331 pKa = 4.37
Molecular weight: 37.32 kDa
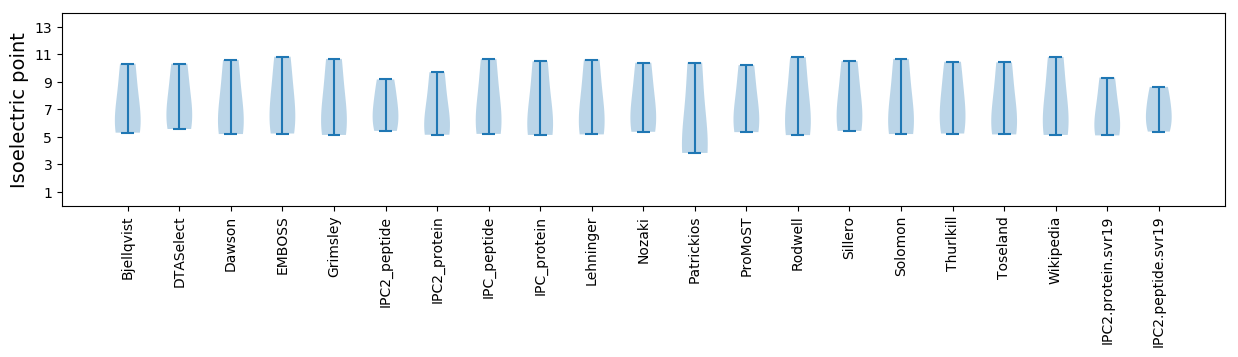
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161J5N3|A0A161J5N3_9VIRU Capsid protein OS=Sewage associated gemycircularvirus 3 OX=1843761 PE=4 SV=1
MM1 pKa = 7.4VYY3 pKa = 10.29RR4 pKa = 11.84RR5 pKa = 11.84LSRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.1SAYY13 pKa = 9.2RR14 pKa = 11.84PRR16 pKa = 11.84SAKK19 pKa = 9.75SRR21 pKa = 11.84YY22 pKa = 7.34GRR24 pKa = 11.84KK25 pKa = 7.78RR26 pKa = 11.84TRR28 pKa = 11.84STRR31 pKa = 11.84VRR33 pKa = 11.84VSRR36 pKa = 11.84RR37 pKa = 11.84MRR39 pKa = 11.84PMSTRR44 pKa = 11.84SLLNRR49 pKa = 11.84TSQKK53 pKa = 10.39KK54 pKa = 8.85RR55 pKa = 11.84DD56 pKa = 3.67NMLSYY61 pKa = 11.09TNTTAADD68 pKa = 3.86PFSTVYY74 pKa = 10.47SATGAVMRR82 pKa = 11.84FPVGQINPGEE92 pKa = 3.96EE93 pKa = 4.56FFYY96 pKa = 10.95VWNATARR103 pKa = 11.84PGEE106 pKa = 4.09TSEE109 pKa = 4.12GQRR112 pKa = 11.84GSKK115 pKa = 9.73IDD117 pKa = 3.35TSLRR121 pKa = 11.84TSSTIYY127 pKa = 10.87AKK129 pKa = 10.39GLRR132 pKa = 11.84EE133 pKa = 4.36KK134 pKa = 9.95ITVEE138 pKa = 4.44TNNSAPWEE146 pKa = 3.86WRR148 pKa = 11.84RR149 pKa = 11.84ICFTSKK155 pKa = 11.11DD156 pKa = 3.86DD157 pKa = 4.07FGEE160 pKa = 4.4RR161 pKa = 11.84DD162 pKa = 3.46PASSTFFRR170 pKa = 11.84QTSSGMVRR178 pKa = 11.84MLSALSTGIYY188 pKa = 10.09LQDD191 pKa = 3.53EE192 pKa = 4.67LFEE195 pKa = 4.39GARR198 pKa = 11.84NVDD201 pKa = 3.21WLSVFTAPLSRR212 pKa = 11.84KK213 pKa = 9.23NFSIKK218 pKa = 9.97YY219 pKa = 9.43DD220 pKa = 3.31RR221 pKa = 11.84TRR223 pKa = 11.84TIRR226 pKa = 11.84STNNSGTIRR235 pKa = 11.84NYY237 pKa = 10.79KK238 pKa = 8.98LWHH241 pKa = 5.96PMEE244 pKa = 5.71HH245 pKa = 5.94NLAYY249 pKa = 10.28QEE251 pKa = 4.22EE252 pKa = 4.64QVGEE256 pKa = 4.29SMTDD260 pKa = 3.07QSVSVTGRR268 pKa = 11.84VGMGNYY274 pKa = 9.59YY275 pKa = 10.35VIDD278 pKa = 3.74MFRR281 pKa = 11.84KK282 pKa = 9.93HH283 pKa = 6.33GNNDD287 pKa = 3.17NDD289 pKa = 3.72STLTFTPEE297 pKa = 3.46ATFFWHH303 pKa = 6.68EE304 pKa = 4.05KK305 pKa = 9.79
MM1 pKa = 7.4VYY3 pKa = 10.29RR4 pKa = 11.84RR5 pKa = 11.84LSRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.1SAYY13 pKa = 9.2RR14 pKa = 11.84PRR16 pKa = 11.84SAKK19 pKa = 9.75SRR21 pKa = 11.84YY22 pKa = 7.34GRR24 pKa = 11.84KK25 pKa = 7.78RR26 pKa = 11.84TRR28 pKa = 11.84STRR31 pKa = 11.84VRR33 pKa = 11.84VSRR36 pKa = 11.84RR37 pKa = 11.84MRR39 pKa = 11.84PMSTRR44 pKa = 11.84SLLNRR49 pKa = 11.84TSQKK53 pKa = 10.39KK54 pKa = 8.85RR55 pKa = 11.84DD56 pKa = 3.67NMLSYY61 pKa = 11.09TNTTAADD68 pKa = 3.86PFSTVYY74 pKa = 10.47SATGAVMRR82 pKa = 11.84FPVGQINPGEE92 pKa = 3.96EE93 pKa = 4.56FFYY96 pKa = 10.95VWNATARR103 pKa = 11.84PGEE106 pKa = 4.09TSEE109 pKa = 4.12GQRR112 pKa = 11.84GSKK115 pKa = 9.73IDD117 pKa = 3.35TSLRR121 pKa = 11.84TSSTIYY127 pKa = 10.87AKK129 pKa = 10.39GLRR132 pKa = 11.84EE133 pKa = 4.36KK134 pKa = 9.95ITVEE138 pKa = 4.44TNNSAPWEE146 pKa = 3.86WRR148 pKa = 11.84RR149 pKa = 11.84ICFTSKK155 pKa = 11.11DD156 pKa = 3.86DD157 pKa = 4.07FGEE160 pKa = 4.4RR161 pKa = 11.84DD162 pKa = 3.46PASSTFFRR170 pKa = 11.84QTSSGMVRR178 pKa = 11.84MLSALSTGIYY188 pKa = 10.09LQDD191 pKa = 3.53EE192 pKa = 4.67LFEE195 pKa = 4.39GARR198 pKa = 11.84NVDD201 pKa = 3.21WLSVFTAPLSRR212 pKa = 11.84KK213 pKa = 9.23NFSIKK218 pKa = 9.97YY219 pKa = 9.43DD220 pKa = 3.31RR221 pKa = 11.84TRR223 pKa = 11.84TIRR226 pKa = 11.84STNNSGTIRR235 pKa = 11.84NYY237 pKa = 10.79KK238 pKa = 8.98LWHH241 pKa = 5.96PMEE244 pKa = 5.71HH245 pKa = 5.94NLAYY249 pKa = 10.28QEE251 pKa = 4.22EE252 pKa = 4.64QVGEE256 pKa = 4.29SMTDD260 pKa = 3.07QSVSVTGRR268 pKa = 11.84VGMGNYY274 pKa = 9.59YY275 pKa = 10.35VIDD278 pKa = 3.74MFRR281 pKa = 11.84KK282 pKa = 9.93HH283 pKa = 6.33GNNDD287 pKa = 3.17NDD289 pKa = 3.72STLTFTPEE297 pKa = 3.46ATFFWHH303 pKa = 6.68EE304 pKa = 4.05KK305 pKa = 9.79
Molecular weight: 35.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
877 |
241 |
331 |
292.3 |
33.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.184 ± 0.877 | 1.71 ± 0.603 |
7.412 ± 1.374 | 5.245 ± 0.111 |
5.587 ± 0.295 | 8.438 ± 1.128 |
2.395 ± 0.471 | 3.649 ± 0.209 |
3.763 ± 0.512 | 7.526 ± 1.155 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.509 ± 0.48 | 3.079 ± 1.089 |
3.649 ± 0.202 | 2.395 ± 0.132 |
9.692 ± 1.168 | 9.008 ± 1.053 |
5.245 ± 2.145 | 5.473 ± 0.137 |
2.052 ± 0.186 | 3.991 ± 0.417 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
