
Felis catus papillomavirus type 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
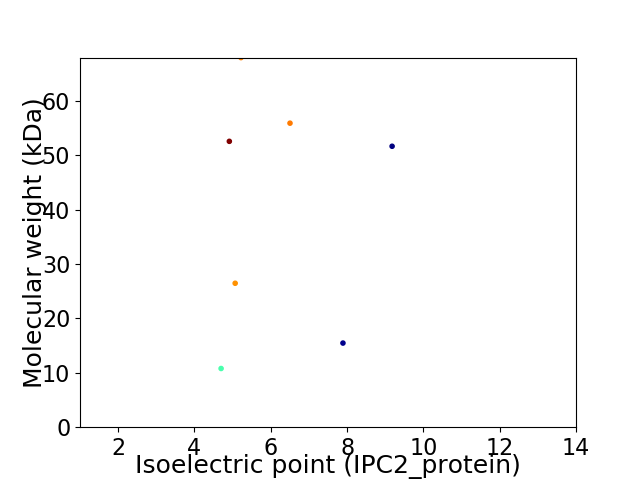
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
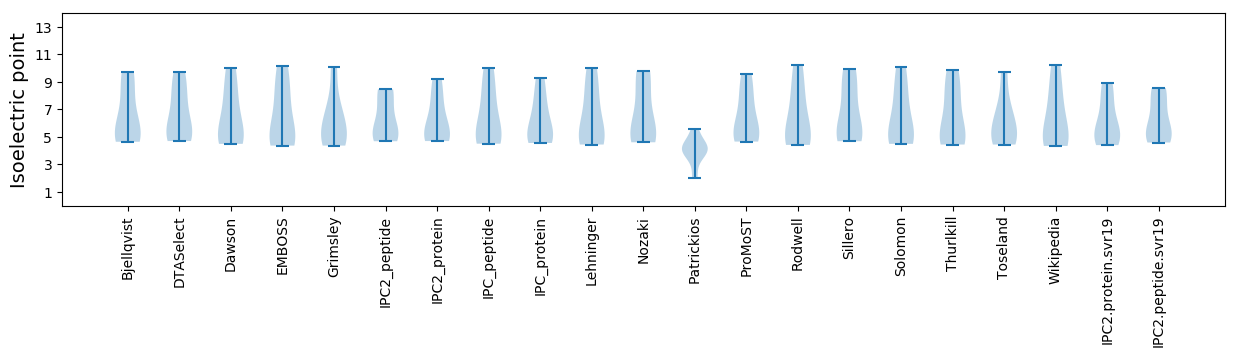
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A223FRE3|A0A223FRE3_9PAPI Minor capsid protein L2 OS=Felis catus papillomavirus type 5 OX=2025339 GN=L2 PE=3 SV=1
MM1 pKa = 7.61IGNIVNIKK9 pKa = 10.33DD10 pKa = 3.44IEE12 pKa = 5.06LDD14 pKa = 3.59LQEE17 pKa = 5.23LVLPANLLAEE27 pKa = 4.46DD28 pKa = 4.26EE29 pKa = 4.96EE30 pKa = 4.53IVEE33 pKa = 4.41AEE35 pKa = 4.13EE36 pKa = 4.33VEE38 pKa = 4.38PPGNSYY44 pKa = 10.77RR45 pKa = 11.84IVTCCVSCHH54 pKa = 4.26STLRR58 pKa = 11.84LWVFATSDD66 pKa = 3.45QIRR69 pKa = 11.84VQQEE73 pKa = 3.74LLLAGLGIICPGCAKK88 pKa = 9.83SHH90 pKa = 5.64CHH92 pKa = 6.61HH93 pKa = 6.92GGEE96 pKa = 4.5QRR98 pKa = 3.59
MM1 pKa = 7.61IGNIVNIKK9 pKa = 10.33DD10 pKa = 3.44IEE12 pKa = 5.06LDD14 pKa = 3.59LQEE17 pKa = 5.23LVLPANLLAEE27 pKa = 4.46DD28 pKa = 4.26EE29 pKa = 4.96EE30 pKa = 4.53IVEE33 pKa = 4.41AEE35 pKa = 4.13EE36 pKa = 4.33VEE38 pKa = 4.38PPGNSYY44 pKa = 10.77RR45 pKa = 11.84IVTCCVSCHH54 pKa = 4.26STLRR58 pKa = 11.84LWVFATSDD66 pKa = 3.45QIRR69 pKa = 11.84VQQEE73 pKa = 3.74LLLAGLGIICPGCAKK88 pKa = 9.83SHH90 pKa = 5.64CHH92 pKa = 6.61HH93 pKa = 6.92GGEE96 pKa = 4.5QRR98 pKa = 3.59
Molecular weight: 10.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223FRD6|A0A223FRD6_9PAPI Protein E6 OS=Felis catus papillomavirus type 5 OX=2025339 GN=E6 PE=3 SV=1
MM1 pKa = 7.59EE2 pKa = 5.19NLNAKK7 pKa = 10.25HH8 pKa = 6.19EE9 pKa = 4.24VLQQVLLAHH18 pKa = 5.64YY19 pKa = 8.93EE20 pKa = 4.03KK21 pKa = 10.77DD22 pKa = 3.49SSKK25 pKa = 10.19LTDD28 pKa = 3.59QIAYY32 pKa = 7.56WEE34 pKa = 4.09LMRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.46SVLHH43 pKa = 6.31HH44 pKa = 6.39YY45 pKa = 9.22ARR47 pKa = 11.84QMGIGTLGFTVVPTLQVSEE66 pKa = 4.44NRR68 pKa = 11.84AKK70 pKa = 10.69SAIKK74 pKa = 9.04MQLLLQSLLNSPFANEE90 pKa = 3.87PWSMRR95 pKa = 11.84DD96 pKa = 3.29TSIEE100 pKa = 3.99RR101 pKa = 11.84LDD103 pKa = 4.32APPKK107 pKa = 10.35NLFKK111 pKa = 10.69KK112 pKa = 10.45RR113 pKa = 11.84PVTVEE118 pKa = 3.91VYY120 pKa = 10.64FDD122 pKa = 4.01NNPDD126 pKa = 3.08NMFPYY131 pKa = 10.27TSWTDD136 pKa = 2.54IYY138 pKa = 11.37YY139 pKa = 10.35QDD141 pKa = 6.04AEE143 pKa = 4.86DD144 pKa = 3.61NWKK147 pKa = 8.55KK148 pKa = 9.34TSGQVDD154 pKa = 3.61NDD156 pKa = 3.22GLYY159 pKa = 10.51YY160 pKa = 10.52VDD162 pKa = 5.76DD163 pKa = 4.65EE164 pKa = 5.53GEE166 pKa = 3.88VHH168 pKa = 6.64YY169 pKa = 10.3YY170 pKa = 9.57VQFGVDD176 pKa = 3.21APVYY180 pKa = 10.16GVTGLWRR187 pKa = 11.84VSYY190 pKa = 10.11KK191 pKa = 10.89NKK193 pKa = 10.21IISTPVSSSSPEE205 pKa = 3.81VQLGQDD211 pKa = 3.23FHH213 pKa = 7.29EE214 pKa = 4.63PQQWYY219 pKa = 8.88PRR221 pKa = 11.84VSVLGFSPASDD232 pKa = 3.19WADD235 pKa = 3.27SPTSTTGTTGSQPLHH250 pKa = 6.71SPISIGSSQSRR261 pKa = 11.84RR262 pKa = 11.84GLRR265 pKa = 11.84GSSRR269 pKa = 11.84GKK271 pKa = 9.56RR272 pKa = 11.84RR273 pKa = 11.84GRR275 pKa = 11.84RR276 pKa = 11.84LSGSPGRR283 pKa = 11.84QGDD286 pKa = 3.65SRR288 pKa = 11.84RR289 pKa = 11.84GRR291 pKa = 11.84GGHH294 pKa = 4.83RR295 pKa = 11.84QEE297 pKa = 4.53SRR299 pKa = 11.84WEE301 pKa = 4.32GSTSPASSSSSLPEE315 pKa = 3.64LSFIGQQRR323 pKa = 11.84GKK325 pKa = 10.47GGRR328 pKa = 11.84GYY330 pKa = 10.43RR331 pKa = 11.84GRR333 pKa = 11.84GRR335 pKa = 11.84GRR337 pKa = 11.84GRR339 pKa = 11.84ASGRR343 pKa = 11.84VPPRR347 pKa = 11.84LSPHH351 pKa = 5.73VSIEE355 pKa = 4.09TLGAGLDD362 pKa = 3.74QPGKK366 pKa = 9.81RR367 pKa = 11.84GGGRR371 pKa = 11.84SEE373 pKa = 3.7EE374 pKa = 4.36AEE376 pKa = 4.05AKK378 pKa = 10.56VGVPVILLKK387 pKa = 11.18GPGNALKK394 pKa = 10.36CWRR397 pKa = 11.84ARR399 pKa = 11.84AKK401 pKa = 9.79QRR403 pKa = 11.84HH404 pKa = 5.7RR405 pKa = 11.84DD406 pKa = 3.41LFGAISTVFSWINKK420 pKa = 8.75CDD422 pKa = 3.43CTRR425 pKa = 11.84IGRR428 pKa = 11.84HH429 pKa = 4.81RR430 pKa = 11.84LVVGFMSEE438 pKa = 4.08GQRR441 pKa = 11.84EE442 pKa = 4.24DD443 pKa = 3.42FLKK446 pKa = 10.7QVTLPGGVDD455 pKa = 2.88WSYY458 pKa = 10.85GTFNSLL464 pKa = 3.9
MM1 pKa = 7.59EE2 pKa = 5.19NLNAKK7 pKa = 10.25HH8 pKa = 6.19EE9 pKa = 4.24VLQQVLLAHH18 pKa = 5.64YY19 pKa = 8.93EE20 pKa = 4.03KK21 pKa = 10.77DD22 pKa = 3.49SSKK25 pKa = 10.19LTDD28 pKa = 3.59QIAYY32 pKa = 7.56WEE34 pKa = 4.09LMRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.46SVLHH43 pKa = 6.31HH44 pKa = 6.39YY45 pKa = 9.22ARR47 pKa = 11.84QMGIGTLGFTVVPTLQVSEE66 pKa = 4.44NRR68 pKa = 11.84AKK70 pKa = 10.69SAIKK74 pKa = 9.04MQLLLQSLLNSPFANEE90 pKa = 3.87PWSMRR95 pKa = 11.84DD96 pKa = 3.29TSIEE100 pKa = 3.99RR101 pKa = 11.84LDD103 pKa = 4.32APPKK107 pKa = 10.35NLFKK111 pKa = 10.69KK112 pKa = 10.45RR113 pKa = 11.84PVTVEE118 pKa = 3.91VYY120 pKa = 10.64FDD122 pKa = 4.01NNPDD126 pKa = 3.08NMFPYY131 pKa = 10.27TSWTDD136 pKa = 2.54IYY138 pKa = 11.37YY139 pKa = 10.35QDD141 pKa = 6.04AEE143 pKa = 4.86DD144 pKa = 3.61NWKK147 pKa = 8.55KK148 pKa = 9.34TSGQVDD154 pKa = 3.61NDD156 pKa = 3.22GLYY159 pKa = 10.51YY160 pKa = 10.52VDD162 pKa = 5.76DD163 pKa = 4.65EE164 pKa = 5.53GEE166 pKa = 3.88VHH168 pKa = 6.64YY169 pKa = 10.3YY170 pKa = 9.57VQFGVDD176 pKa = 3.21APVYY180 pKa = 10.16GVTGLWRR187 pKa = 11.84VSYY190 pKa = 10.11KK191 pKa = 10.89NKK193 pKa = 10.21IISTPVSSSSPEE205 pKa = 3.81VQLGQDD211 pKa = 3.23FHH213 pKa = 7.29EE214 pKa = 4.63PQQWYY219 pKa = 8.88PRR221 pKa = 11.84VSVLGFSPASDD232 pKa = 3.19WADD235 pKa = 3.27SPTSTTGTTGSQPLHH250 pKa = 6.71SPISIGSSQSRR261 pKa = 11.84RR262 pKa = 11.84GLRR265 pKa = 11.84GSSRR269 pKa = 11.84GKK271 pKa = 9.56RR272 pKa = 11.84RR273 pKa = 11.84GRR275 pKa = 11.84RR276 pKa = 11.84LSGSPGRR283 pKa = 11.84QGDD286 pKa = 3.65SRR288 pKa = 11.84RR289 pKa = 11.84GRR291 pKa = 11.84GGHH294 pKa = 4.83RR295 pKa = 11.84QEE297 pKa = 4.53SRR299 pKa = 11.84WEE301 pKa = 4.32GSTSPASSSSSLPEE315 pKa = 3.64LSFIGQQRR323 pKa = 11.84GKK325 pKa = 10.47GGRR328 pKa = 11.84GYY330 pKa = 10.43RR331 pKa = 11.84GRR333 pKa = 11.84GRR335 pKa = 11.84GRR337 pKa = 11.84GRR339 pKa = 11.84ASGRR343 pKa = 11.84VPPRR347 pKa = 11.84LSPHH351 pKa = 5.73VSIEE355 pKa = 4.09TLGAGLDD362 pKa = 3.74QPGKK366 pKa = 9.81RR367 pKa = 11.84GGGRR371 pKa = 11.84SEE373 pKa = 3.7EE374 pKa = 4.36AEE376 pKa = 4.05AKK378 pKa = 10.56VGVPVILLKK387 pKa = 11.18GPGNALKK394 pKa = 10.36CWRR397 pKa = 11.84ARR399 pKa = 11.84AKK401 pKa = 9.79QRR403 pKa = 11.84HH404 pKa = 5.7RR405 pKa = 11.84DD406 pKa = 3.41LFGAISTVFSWINKK420 pKa = 8.75CDD422 pKa = 3.43CTRR425 pKa = 11.84IGRR428 pKa = 11.84HH429 pKa = 4.81RR430 pKa = 11.84LVVGFMSEE438 pKa = 4.08GQRR441 pKa = 11.84EE442 pKa = 4.24DD443 pKa = 3.42FLKK446 pKa = 10.7QVTLPGGVDD455 pKa = 2.88WSYY458 pKa = 10.85GTFNSLL464 pKa = 3.9
Molecular weight: 51.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2530 |
98 |
595 |
361.4 |
40.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.625 ± 0.468 | 2.451 ± 0.815 |
5.652 ± 0.382 | 6.838 ± 0.728 |
3.715 ± 0.438 | 9.091 ± 1.171 |
2.253 ± 0.239 | 3.597 ± 0.348 |
4.664 ± 0.582 | 8.972 ± 0.571 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.818 ± 0.367 | 3.636 ± 0.489 |
5.929 ± 0.862 | 4.19 ± 0.397 |
6.482 ± 0.564 | 8.893 ± 0.599 |
5.81 ± 0.605 | 6.996 ± 0.633 |
1.581 ± 0.289 | 2.806 ± 0.365 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
