
Yongsan tombus-like virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; unclassified Tombusviridae
Average proteome isoelectric point is 8.81
Get precalculated fractions of proteins
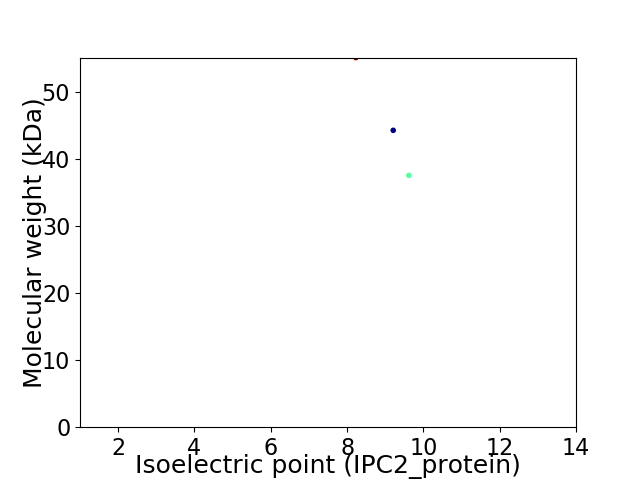
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385FQG9|A0A385FQG9_9TOMB Uncharacterized protein OS=Yongsan tombus-like virus 1 OX=2315813 PE=4 SV=1
MM1 pKa = 7.41SLNDD5 pKa = 3.46KK6 pKa = 9.87VLPGSCLSAGACDD19 pKa = 4.24HH20 pKa = 5.37VCKK23 pKa = 10.49RR24 pKa = 11.84YY25 pKa = 6.35TQQMFTIQGFTSEE38 pKa = 4.12VWTHH42 pKa = 6.14GSCVCNEE49 pKa = 3.94VIALKK54 pKa = 10.37NRR56 pKa = 11.84HH57 pKa = 5.21QLCDD61 pKa = 3.4GSAYY65 pKa = 10.81NSTVDD70 pKa = 4.02LRR72 pKa = 11.84RR73 pKa = 11.84PLRR76 pKa = 11.84KK77 pKa = 9.78LITNLEE83 pKa = 4.29PCSEE87 pKa = 4.25QTVIAHH93 pKa = 7.28ADD95 pKa = 3.5GSRR98 pKa = 11.84KK99 pKa = 9.7KK100 pKa = 10.46LLQRR104 pKa = 11.84AADD107 pKa = 3.68SLSTHH112 pKa = 5.62GLEE115 pKa = 6.11HH116 pKa = 6.74KK117 pKa = 10.28DD118 pKa = 2.94AWVKK122 pKa = 10.38MFLKK126 pKa = 10.25DD127 pKa = 4.24DD128 pKa = 4.11KK129 pKa = 11.43YY130 pKa = 10.51HH131 pKa = 5.34TPEE134 pKa = 4.02YY135 pKa = 9.99KK136 pKa = 10.36APRR139 pKa = 11.84CIQYY143 pKa = 10.18RR144 pKa = 11.84NKK146 pKa = 10.62RR147 pKa = 11.84YY148 pKa = 9.98GLRR151 pKa = 11.84LATFLHH157 pKa = 6.98PIEE160 pKa = 4.32QRR162 pKa = 11.84VITLKK167 pKa = 11.09HH168 pKa = 5.94NGTHH172 pKa = 5.58VFAKK176 pKa = 9.07GRR178 pKa = 11.84NMKK181 pKa = 9.93QRR183 pKa = 11.84GRR185 pKa = 11.84DD186 pKa = 3.19IAAKK190 pKa = 10.39LLPNWVAVSMDD201 pKa = 3.08HH202 pKa = 7.32SKK204 pKa = 10.78FDD206 pKa = 3.49SHH208 pKa = 8.04VNEE211 pKa = 3.89HH212 pKa = 6.67LLRR215 pKa = 11.84LEE217 pKa = 3.5HH218 pKa = 6.94WYY220 pKa = 10.98YY221 pKa = 10.11NQCINDD227 pKa = 4.41PEE229 pKa = 4.66LKK231 pKa = 9.85WLLEE235 pKa = 3.97CQLRR239 pKa = 11.84NKK241 pKa = 10.33GFTKK245 pKa = 10.75NNTRR249 pKa = 11.84YY250 pKa = 5.23TTRR253 pKa = 11.84ATRR256 pKa = 11.84MSGDD260 pKa = 3.33QNTGLGNCIINYY272 pKa = 10.34AMTKK276 pKa = 10.88AMLDD280 pKa = 3.56HH281 pKa = 7.29LDD283 pKa = 3.84IPHH286 pKa = 6.82NLYY289 pKa = 10.06IDD291 pKa = 3.99GDD293 pKa = 4.02DD294 pKa = 4.51FIVFVHH300 pKa = 6.87RR301 pKa = 11.84KK302 pKa = 8.23HH303 pKa = 6.37KK304 pKa = 10.45HH305 pKa = 5.78LIDD308 pKa = 4.4PNWYY312 pKa = 9.1KK313 pKa = 10.65QFGMKK318 pKa = 7.62TTTDD322 pKa = 3.26QITDD326 pKa = 3.62VLEE329 pKa = 5.48HH330 pKa = 7.39IDD332 pKa = 3.8FCQCRR337 pKa = 11.84PVYY340 pKa = 10.9DD341 pKa = 3.96GDD343 pKa = 4.0SYY345 pKa = 11.56TLVRR349 pKa = 11.84NPARR353 pKa = 11.84LLARR357 pKa = 11.84LPWLVGPINGRR368 pKa = 11.84SPWNIVASAAQCEE381 pKa = 4.11ISLGYY386 pKa = 10.06GLPIGQYY393 pKa = 8.61VGHH396 pKa = 6.44NVFLYY401 pKa = 10.7ASNKK405 pKa = 9.28GGRR408 pKa = 11.84FKK410 pKa = 11.27SNCVAEE416 pKa = 4.66WRR418 pKa = 11.84HH419 pKa = 5.18KK420 pKa = 8.27MEE422 pKa = 4.7RR423 pKa = 11.84MKK425 pKa = 10.84AGNLKK430 pKa = 10.22PVDD433 pKa = 3.77CAPGVRR439 pKa = 11.84ASYY442 pKa = 9.63EE443 pKa = 4.13RR444 pKa = 11.84AWGISIPQQYY454 pKa = 9.78IIEE457 pKa = 4.62HH458 pKa = 5.85ATIAPPFEE466 pKa = 4.69EE467 pKa = 4.98EE468 pKa = 3.76WDD470 pKa = 3.5HH471 pKa = 6.74TPFDD475 pKa = 4.04RR476 pKa = 11.84KK477 pKa = 8.86TYY479 pKa = 10.53
MM1 pKa = 7.41SLNDD5 pKa = 3.46KK6 pKa = 9.87VLPGSCLSAGACDD19 pKa = 4.24HH20 pKa = 5.37VCKK23 pKa = 10.49RR24 pKa = 11.84YY25 pKa = 6.35TQQMFTIQGFTSEE38 pKa = 4.12VWTHH42 pKa = 6.14GSCVCNEE49 pKa = 3.94VIALKK54 pKa = 10.37NRR56 pKa = 11.84HH57 pKa = 5.21QLCDD61 pKa = 3.4GSAYY65 pKa = 10.81NSTVDD70 pKa = 4.02LRR72 pKa = 11.84RR73 pKa = 11.84PLRR76 pKa = 11.84KK77 pKa = 9.78LITNLEE83 pKa = 4.29PCSEE87 pKa = 4.25QTVIAHH93 pKa = 7.28ADD95 pKa = 3.5GSRR98 pKa = 11.84KK99 pKa = 9.7KK100 pKa = 10.46LLQRR104 pKa = 11.84AADD107 pKa = 3.68SLSTHH112 pKa = 5.62GLEE115 pKa = 6.11HH116 pKa = 6.74KK117 pKa = 10.28DD118 pKa = 2.94AWVKK122 pKa = 10.38MFLKK126 pKa = 10.25DD127 pKa = 4.24DD128 pKa = 4.11KK129 pKa = 11.43YY130 pKa = 10.51HH131 pKa = 5.34TPEE134 pKa = 4.02YY135 pKa = 9.99KK136 pKa = 10.36APRR139 pKa = 11.84CIQYY143 pKa = 10.18RR144 pKa = 11.84NKK146 pKa = 10.62RR147 pKa = 11.84YY148 pKa = 9.98GLRR151 pKa = 11.84LATFLHH157 pKa = 6.98PIEE160 pKa = 4.32QRR162 pKa = 11.84VITLKK167 pKa = 11.09HH168 pKa = 5.94NGTHH172 pKa = 5.58VFAKK176 pKa = 9.07GRR178 pKa = 11.84NMKK181 pKa = 9.93QRR183 pKa = 11.84GRR185 pKa = 11.84DD186 pKa = 3.19IAAKK190 pKa = 10.39LLPNWVAVSMDD201 pKa = 3.08HH202 pKa = 7.32SKK204 pKa = 10.78FDD206 pKa = 3.49SHH208 pKa = 8.04VNEE211 pKa = 3.89HH212 pKa = 6.67LLRR215 pKa = 11.84LEE217 pKa = 3.5HH218 pKa = 6.94WYY220 pKa = 10.98YY221 pKa = 10.11NQCINDD227 pKa = 4.41PEE229 pKa = 4.66LKK231 pKa = 9.85WLLEE235 pKa = 3.97CQLRR239 pKa = 11.84NKK241 pKa = 10.33GFTKK245 pKa = 10.75NNTRR249 pKa = 11.84YY250 pKa = 5.23TTRR253 pKa = 11.84ATRR256 pKa = 11.84MSGDD260 pKa = 3.33QNTGLGNCIINYY272 pKa = 10.34AMTKK276 pKa = 10.88AMLDD280 pKa = 3.56HH281 pKa = 7.29LDD283 pKa = 3.84IPHH286 pKa = 6.82NLYY289 pKa = 10.06IDD291 pKa = 3.99GDD293 pKa = 4.02DD294 pKa = 4.51FIVFVHH300 pKa = 6.87RR301 pKa = 11.84KK302 pKa = 8.23HH303 pKa = 6.37KK304 pKa = 10.45HH305 pKa = 5.78LIDD308 pKa = 4.4PNWYY312 pKa = 9.1KK313 pKa = 10.65QFGMKK318 pKa = 7.62TTTDD322 pKa = 3.26QITDD326 pKa = 3.62VLEE329 pKa = 5.48HH330 pKa = 7.39IDD332 pKa = 3.8FCQCRR337 pKa = 11.84PVYY340 pKa = 10.9DD341 pKa = 3.96GDD343 pKa = 4.0SYY345 pKa = 11.56TLVRR349 pKa = 11.84NPARR353 pKa = 11.84LLARR357 pKa = 11.84LPWLVGPINGRR368 pKa = 11.84SPWNIVASAAQCEE381 pKa = 4.11ISLGYY386 pKa = 10.06GLPIGQYY393 pKa = 8.61VGHH396 pKa = 6.44NVFLYY401 pKa = 10.7ASNKK405 pKa = 9.28GGRR408 pKa = 11.84FKK410 pKa = 11.27SNCVAEE416 pKa = 4.66WRR418 pKa = 11.84HH419 pKa = 5.18KK420 pKa = 8.27MEE422 pKa = 4.7RR423 pKa = 11.84MKK425 pKa = 10.84AGNLKK430 pKa = 10.22PVDD433 pKa = 3.77CAPGVRR439 pKa = 11.84ASYY442 pKa = 9.63EE443 pKa = 4.13RR444 pKa = 11.84AWGISIPQQYY454 pKa = 9.78IIEE457 pKa = 4.62HH458 pKa = 5.85ATIAPPFEE466 pKa = 4.69EE467 pKa = 4.98EE468 pKa = 3.76WDD470 pKa = 3.5HH471 pKa = 6.74TPFDD475 pKa = 4.04RR476 pKa = 11.84KK477 pKa = 8.86TYY479 pKa = 10.53
Molecular weight: 55.11 kDa
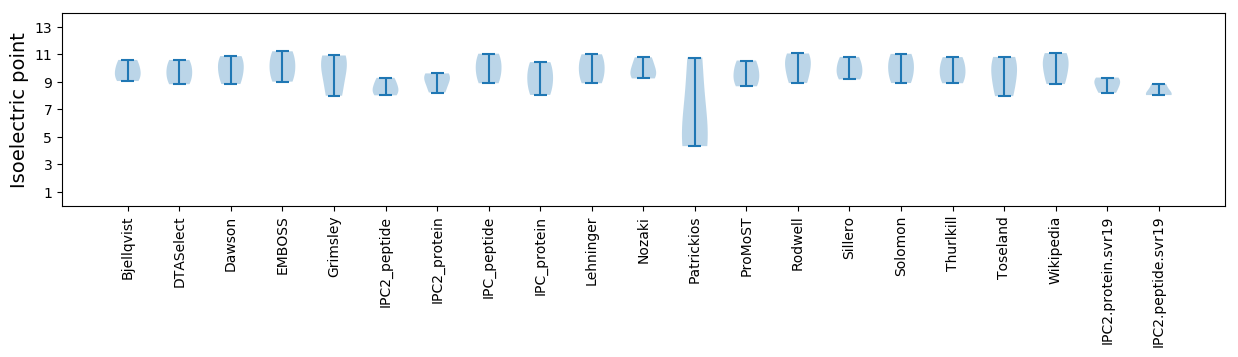
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385FQH1|A0A385FQH1_9TOMB Uncharacterized protein OS=Yongsan tombus-like virus 1 OX=2315813 PE=4 SV=1
MM1 pKa = 7.93RR2 pKa = 11.84ALQVEE7 pKa = 4.48LQHH10 pKa = 6.86LRR12 pKa = 11.84RR13 pKa = 11.84ANSALQARR21 pKa = 11.84VRR23 pKa = 11.84DD24 pKa = 3.88LEE26 pKa = 4.07GLITRR31 pKa = 11.84NTSAPLGGSGPITTIPRR48 pKa = 11.84GSRR51 pKa = 11.84RR52 pKa = 11.84STLVRR57 pKa = 11.84ASGVASTPNLNSSPEE72 pKa = 4.0PRR74 pKa = 11.84SQRR77 pKa = 11.84PVPHH81 pKa = 6.71VNTTPPVSRR90 pKa = 11.84RR91 pKa = 11.84QSSTSPRR98 pKa = 11.84VGTPSRR104 pKa = 11.84PQSVTALTRR113 pKa = 11.84PVTPASAGQAAEE125 pKa = 4.16HH126 pKa = 6.13STVSTQLSSGPALSPAPEE144 pKa = 4.11TTASKK149 pKa = 9.68TVVSHH154 pKa = 6.92PYY156 pKa = 10.19AGAILEE162 pKa = 4.09ATRR165 pKa = 11.84AKK167 pKa = 10.96GEE169 pKa = 4.07LAHH172 pKa = 7.13DD173 pKa = 3.76EE174 pKa = 4.39RR175 pKa = 11.84PVYY178 pKa = 10.39VADD181 pKa = 3.72HH182 pKa = 6.78AIDD185 pKa = 3.56RR186 pKa = 11.84SLRR189 pKa = 11.84EE190 pKa = 3.84SVGITRR196 pKa = 11.84RR197 pKa = 11.84AVNKK201 pKa = 6.42MTAVDD206 pKa = 3.72EE207 pKa = 4.41EE208 pKa = 4.72EE209 pKa = 4.28PKK211 pKa = 11.21GMWRR215 pKa = 11.84YY216 pKa = 8.98LPKK219 pKa = 10.26VRR221 pKa = 11.84SPALRR226 pKa = 11.84ADD228 pKa = 3.42KK229 pKa = 11.05YY230 pKa = 11.73LLFFLLNKK238 pKa = 9.92FAFIEE243 pKa = 4.19RR244 pKa = 11.84SPDD247 pKa = 2.62TMRR250 pKa = 11.84RR251 pKa = 11.84MWEE254 pKa = 3.89SLNQVMRR261 pKa = 11.84DD262 pKa = 2.94FDD264 pKa = 3.9TRR266 pKa = 11.84NNTIEE271 pKa = 4.27EE272 pKa = 4.35IYY274 pKa = 10.41LLKK277 pKa = 10.5HH278 pKa = 4.54QAVRR282 pKa = 11.84AAMVPPAEE290 pKa = 4.27EE291 pKa = 3.69LRR293 pKa = 11.84TRR295 pKa = 11.84KK296 pKa = 9.48LLQEE300 pKa = 4.39APNLDD305 pKa = 3.2KK306 pKa = 10.87HH307 pKa = 7.15AKK309 pKa = 7.03FTKK312 pKa = 10.08DD313 pKa = 2.53GHH315 pKa = 6.69LGHH318 pKa = 7.07KK319 pKa = 10.33LEE321 pKa = 5.29ASLSTIGNLFQPTKK335 pKa = 10.72SLPTKK340 pKa = 10.28KK341 pKa = 10.5
MM1 pKa = 7.93RR2 pKa = 11.84ALQVEE7 pKa = 4.48LQHH10 pKa = 6.86LRR12 pKa = 11.84RR13 pKa = 11.84ANSALQARR21 pKa = 11.84VRR23 pKa = 11.84DD24 pKa = 3.88LEE26 pKa = 4.07GLITRR31 pKa = 11.84NTSAPLGGSGPITTIPRR48 pKa = 11.84GSRR51 pKa = 11.84RR52 pKa = 11.84STLVRR57 pKa = 11.84ASGVASTPNLNSSPEE72 pKa = 4.0PRR74 pKa = 11.84SQRR77 pKa = 11.84PVPHH81 pKa = 6.71VNTTPPVSRR90 pKa = 11.84RR91 pKa = 11.84QSSTSPRR98 pKa = 11.84VGTPSRR104 pKa = 11.84PQSVTALTRR113 pKa = 11.84PVTPASAGQAAEE125 pKa = 4.16HH126 pKa = 6.13STVSTQLSSGPALSPAPEE144 pKa = 4.11TTASKK149 pKa = 9.68TVVSHH154 pKa = 6.92PYY156 pKa = 10.19AGAILEE162 pKa = 4.09ATRR165 pKa = 11.84AKK167 pKa = 10.96GEE169 pKa = 4.07LAHH172 pKa = 7.13DD173 pKa = 3.76EE174 pKa = 4.39RR175 pKa = 11.84PVYY178 pKa = 10.39VADD181 pKa = 3.72HH182 pKa = 6.78AIDD185 pKa = 3.56RR186 pKa = 11.84SLRR189 pKa = 11.84EE190 pKa = 3.84SVGITRR196 pKa = 11.84RR197 pKa = 11.84AVNKK201 pKa = 6.42MTAVDD206 pKa = 3.72EE207 pKa = 4.41EE208 pKa = 4.72EE209 pKa = 4.28PKK211 pKa = 11.21GMWRR215 pKa = 11.84YY216 pKa = 8.98LPKK219 pKa = 10.26VRR221 pKa = 11.84SPALRR226 pKa = 11.84ADD228 pKa = 3.42KK229 pKa = 11.05YY230 pKa = 11.73LLFFLLNKK238 pKa = 9.92FAFIEE243 pKa = 4.19RR244 pKa = 11.84SPDD247 pKa = 2.62TMRR250 pKa = 11.84RR251 pKa = 11.84MWEE254 pKa = 3.89SLNQVMRR261 pKa = 11.84DD262 pKa = 2.94FDD264 pKa = 3.9TRR266 pKa = 11.84NNTIEE271 pKa = 4.27EE272 pKa = 4.35IYY274 pKa = 10.41LLKK277 pKa = 10.5HH278 pKa = 4.54QAVRR282 pKa = 11.84AAMVPPAEE290 pKa = 4.27EE291 pKa = 3.69LRR293 pKa = 11.84TRR295 pKa = 11.84KK296 pKa = 9.48LLQEE300 pKa = 4.39APNLDD305 pKa = 3.2KK306 pKa = 10.87HH307 pKa = 7.15AKK309 pKa = 7.03FTKK312 pKa = 10.08DD313 pKa = 2.53GHH315 pKa = 6.69LGHH318 pKa = 7.07KK319 pKa = 10.33LEE321 pKa = 5.29ASLSTIGNLFQPTKK335 pKa = 10.72SLPTKK340 pKa = 10.28KK341 pKa = 10.5
Molecular weight: 37.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1228 |
341 |
479 |
409.3 |
45.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.062 ± 0.909 | 1.547 ± 0.927 |
3.746 ± 1.058 | 4.56 ± 0.488 |
2.85 ± 0.3 | 5.537 ± 0.35 |
3.094 ± 0.998 | 4.642 ± 0.66 |
4.886 ± 0.818 | 9.365 ± 0.295 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.954 ± 0.2 | 4.479 ± 0.594 |
6.84 ± 1.134 | 4.56 ± 0.611 |
7.166 ± 1.061 | 7.573 ± 1.383 |
8.143 ± 1.288 | 6.596 ± 0.756 |
1.466 ± 0.453 | 2.932 ± 0.705 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
