
Simian torque teno virus 30
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus; Torque teno chlorocebus virus 1
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
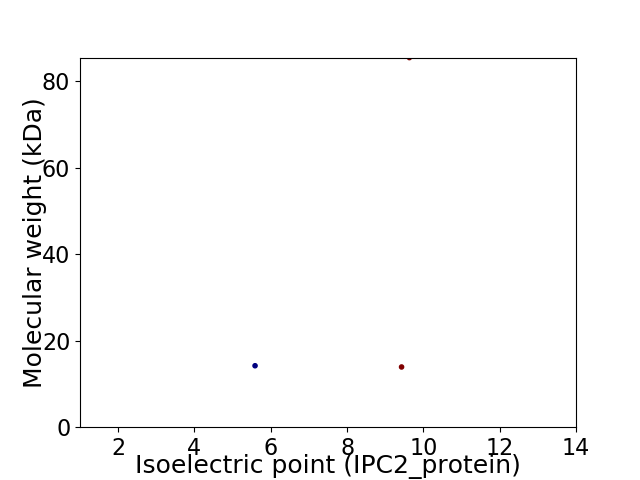
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
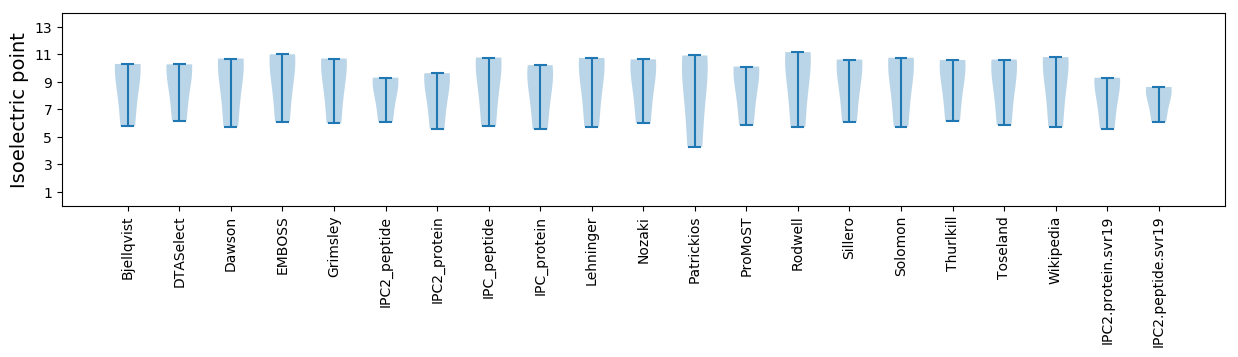
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IMW1|A0A0C5IMW1_9VIRU ORF2 OS=Simian torque teno virus 30 OX=1619218 PE=4 SV=1
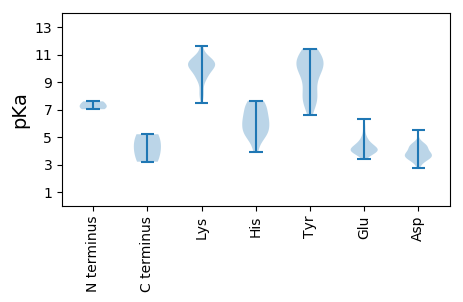
MM1 pKa = 7.02YY2 pKa = 10.15FRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.47AKK11 pKa = 9.86LGSVGDD17 pKa = 4.08YY18 pKa = 11.3LEE20 pKa = 5.19LSHH23 pKa = 7.37PGLPTGTPRR32 pKa = 11.84AMAKK36 pKa = 9.96RR37 pKa = 11.84PRR39 pKa = 11.84EE40 pKa = 4.13TYY42 pKa = 7.36EE43 pKa = 4.07TTSKK47 pKa = 10.64EE48 pKa = 3.86AATWLHH54 pKa = 6.13LVGEE58 pKa = 4.68THH60 pKa = 7.5DD61 pKa = 3.84LWCPCGKK68 pKa = 9.54PLPHH72 pKa = 7.13LSALLNTVGAATTTSTCRR90 pKa = 11.84TGGEE94 pKa = 4.0DD95 pKa = 3.1AAGGGRR101 pKa = 11.84AAGAGPATEE110 pKa = 6.31GGDD113 pKa = 3.19GWDD116 pKa = 5.5DD117 pKa = 3.54INLDD121 pKa = 3.71EE122 pKa = 5.6LLADD126 pKa = 4.24VAAEE130 pKa = 3.9DD131 pKa = 4.34AEE133 pKa = 4.34RR134 pKa = 5.2
MM1 pKa = 7.02YY2 pKa = 10.15FRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.47AKK11 pKa = 9.86LGSVGDD17 pKa = 4.08YY18 pKa = 11.3LEE20 pKa = 5.19LSHH23 pKa = 7.37PGLPTGTPRR32 pKa = 11.84AMAKK36 pKa = 9.96RR37 pKa = 11.84PRR39 pKa = 11.84EE40 pKa = 4.13TYY42 pKa = 7.36EE43 pKa = 4.07TTSKK47 pKa = 10.64EE48 pKa = 3.86AATWLHH54 pKa = 6.13LVGEE58 pKa = 4.68THH60 pKa = 7.5DD61 pKa = 3.84LWCPCGKK68 pKa = 9.54PLPHH72 pKa = 7.13LSALLNTVGAATTTSTCRR90 pKa = 11.84TGGEE94 pKa = 4.0DD95 pKa = 3.1AAGGGRR101 pKa = 11.84AAGAGPATEE110 pKa = 6.31GGDD113 pKa = 3.19GWDD116 pKa = 5.5DD117 pKa = 3.54INLDD121 pKa = 3.71EE122 pKa = 5.6LLADD126 pKa = 4.24VAAEE130 pKa = 3.9DD131 pKa = 4.34AEE133 pKa = 4.34RR134 pKa = 5.2
Molecular weight: 14.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IMW1|A0A0C5IMW1_9VIRU ORF2 OS=Simian torque teno virus 30 OX=1619218 PE=4 SV=1
MM1 pKa = 7.6PYY3 pKa = 7.68WWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84WPRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84WTRR20 pKa = 11.84YY21 pKa = 7.92RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84WMGRR29 pKa = 11.84YY30 pKa = 8.84KK31 pKa = 10.3PRR33 pKa = 11.84RR34 pKa = 11.84ATRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84TVRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84FSGYY54 pKa = 6.63TRR56 pKa = 11.84RR57 pKa = 11.84ATRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84GRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84LTVTLKK74 pKa = 9.2QWKK77 pKa = 9.32PEE79 pKa = 3.96TLRR82 pKa = 11.84SLRR85 pKa = 11.84VTGLFPLIVCGSGASRR101 pKa = 11.84NNWIQRR107 pKa = 11.84SFDD110 pKa = 3.43VPGPDD115 pKa = 3.43TPIGGNLSVSVWTMRR130 pKa = 11.84VLYY133 pKa = 9.83EE134 pKa = 3.58EE135 pKa = 4.5WQLHH139 pKa = 5.24RR140 pKa = 11.84NNWSRR145 pKa = 11.84TNSDD149 pKa = 3.65LDD151 pKa = 3.42LVMYY155 pKa = 7.81YY156 pKa = 11.04GCTLKK161 pKa = 10.52FWRR164 pKa = 11.84HH165 pKa = 5.74PYY167 pKa = 9.55VDD169 pKa = 4.46YY170 pKa = 10.37IVKK173 pKa = 10.44YY174 pKa = 10.42IRR176 pKa = 11.84NTPFKK181 pKa = 10.48TEE183 pKa = 4.07EE184 pKa = 3.87LTHH187 pKa = 6.43AAGHH191 pKa = 6.21PMVLMLSKK199 pKa = 10.29HH200 pKa = 5.42HH201 pKa = 6.31VVVPSLLTKK210 pKa = 8.84PHH212 pKa = 4.8GRR214 pKa = 11.84RR215 pKa = 11.84WIKK218 pKa = 9.24VTVRR222 pKa = 11.84PPRR225 pKa = 11.84MLTSKK230 pKa = 10.56FYY232 pKa = 10.56FQSDD236 pKa = 3.69FCNVNLLGIVASACKK251 pKa = 10.34LNSPWLEE258 pKa = 3.73PDD260 pKa = 4.28KK261 pKa = 10.48ITPCITFYY269 pKa = 11.44ALKK272 pKa = 9.24NTVFKK277 pKa = 10.78NQNITEE283 pKa = 4.13AQNAYY288 pKa = 7.72TALTAIIVQNKK299 pKa = 7.48EE300 pKa = 3.39MAYY303 pKa = 8.44NTWLEE308 pKa = 4.15HH309 pKa = 5.51HH310 pKa = 6.85WGSFGNGKK318 pKa = 9.87AFNITNVKK326 pKa = 10.44DD327 pKa = 3.79NSTLTQNQINTLRR340 pKa = 11.84QTAEE344 pKa = 3.64NTFKK348 pKa = 10.91SRR350 pKa = 11.84YY351 pKa = 7.57NFIYY355 pKa = 9.86EE356 pKa = 4.2AQPPTTFTYY365 pKa = 11.11YY366 pKa = 10.88NLHH369 pKa = 5.31GWGLYY374 pKa = 10.02SPLWLDD380 pKa = 3.42PDD382 pKa = 4.21RR383 pKa = 11.84IEE385 pKa = 4.92PEE387 pKa = 3.79SEE389 pKa = 3.72AAFLAVRR396 pKa = 11.84YY397 pKa = 9.9NPLNDD402 pKa = 3.56KK403 pKa = 11.64GEE405 pKa = 4.47GNWVALQPLTKK416 pKa = 9.87EE417 pKa = 4.5LPILDD422 pKa = 4.53DD423 pKa = 3.51TCKK426 pKa = 10.0MVLKK430 pKa = 10.28DD431 pKa = 3.7YY432 pKa = 10.21PLWLILYY439 pKa = 7.08GYY441 pKa = 9.01PDD443 pKa = 4.42AATKK447 pKa = 10.12ILSGSSPLNNTRR459 pKa = 11.84LIIRR463 pKa = 11.84CQYY466 pKa = 9.6TDD468 pKa = 3.88PPLAFPQATDD478 pKa = 3.25YY479 pKa = 11.22KK480 pKa = 10.01RR481 pKa = 11.84GYY483 pKa = 8.43TIYY486 pKa = 10.83SRR488 pKa = 11.84DD489 pKa = 3.43FATGRR494 pKa = 11.84MPGGNTFIPITTARR508 pKa = 11.84LWYY511 pKa = 9.44PRR513 pKa = 11.84LQNQLDD519 pKa = 3.85VVEE522 pKa = 5.8AIVNCGPLMPRR533 pKa = 11.84DD534 pKa = 4.4DD535 pKa = 4.7LHH537 pKa = 7.64QGWAVTMGYY546 pKa = 10.29KK547 pKa = 10.22YY548 pKa = 10.47RR549 pKa = 11.84FQLGGNLPPKK559 pKa = 9.87QDD561 pKa = 3.49PVDD564 pKa = 3.64PCKK567 pKa = 10.74VPKK570 pKa = 10.29RR571 pKa = 11.84EE572 pKa = 4.22LPDD575 pKa = 3.8PSAEE579 pKa = 3.95LTAVQIVDD587 pKa = 4.16PATMDD592 pKa = 4.0PLSTLHH598 pKa = 6.26PWDD601 pKa = 3.77YY602 pKa = 10.46RR603 pKa = 11.84RR604 pKa = 11.84SMLTSSGIKK613 pKa = 10.02RR614 pKa = 11.84MSADD618 pKa = 3.81RR619 pKa = 11.84EE620 pKa = 4.43TDD622 pKa = 2.98SSLYY626 pKa = 10.26PGATGVAKK634 pKa = 10.38RR635 pKa = 11.84PKK637 pKa = 9.92TDD639 pKa = 2.72VPTQKK644 pKa = 10.73GEE646 pKa = 4.03EE647 pKa = 4.16PGPGEE652 pKa = 4.05RR653 pKa = 11.84LRR655 pKa = 11.84EE656 pKa = 3.99VLQSLLEE663 pKa = 4.07EE664 pKa = 4.28QQQEE668 pKa = 4.6TEE670 pKa = 4.97DD671 pKa = 4.32PPSPPPGEE679 pKa = 4.49RR680 pKa = 11.84VQQPEE685 pKa = 4.52LFQQLALRR693 pKa = 11.84YY694 pKa = 8.85QLQQQRR700 pKa = 11.84EE701 pKa = 4.28RR702 pKa = 11.84QGKK705 pKa = 8.09LAQGIKK711 pKa = 11.01YY712 pKa = 7.92MFQQLIKK719 pKa = 8.17TQQGVQVDD727 pKa = 3.9PRR729 pKa = 11.84LLL731 pKa = 4.26
MM1 pKa = 7.6PYY3 pKa = 7.68WWRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84WPRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84WTRR20 pKa = 11.84YY21 pKa = 7.92RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84WMGRR29 pKa = 11.84YY30 pKa = 8.84KK31 pKa = 10.3PRR33 pKa = 11.84RR34 pKa = 11.84ATRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84TVRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84FSGYY54 pKa = 6.63TRR56 pKa = 11.84RR57 pKa = 11.84ATRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84GRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84LTVTLKK74 pKa = 9.2QWKK77 pKa = 9.32PEE79 pKa = 3.96TLRR82 pKa = 11.84SLRR85 pKa = 11.84VTGLFPLIVCGSGASRR101 pKa = 11.84NNWIQRR107 pKa = 11.84SFDD110 pKa = 3.43VPGPDD115 pKa = 3.43TPIGGNLSVSVWTMRR130 pKa = 11.84VLYY133 pKa = 9.83EE134 pKa = 3.58EE135 pKa = 4.5WQLHH139 pKa = 5.24RR140 pKa = 11.84NNWSRR145 pKa = 11.84TNSDD149 pKa = 3.65LDD151 pKa = 3.42LVMYY155 pKa = 7.81YY156 pKa = 11.04GCTLKK161 pKa = 10.52FWRR164 pKa = 11.84HH165 pKa = 5.74PYY167 pKa = 9.55VDD169 pKa = 4.46YY170 pKa = 10.37IVKK173 pKa = 10.44YY174 pKa = 10.42IRR176 pKa = 11.84NTPFKK181 pKa = 10.48TEE183 pKa = 4.07EE184 pKa = 3.87LTHH187 pKa = 6.43AAGHH191 pKa = 6.21PMVLMLSKK199 pKa = 10.29HH200 pKa = 5.42HH201 pKa = 6.31VVVPSLLTKK210 pKa = 8.84PHH212 pKa = 4.8GRR214 pKa = 11.84RR215 pKa = 11.84WIKK218 pKa = 9.24VTVRR222 pKa = 11.84PPRR225 pKa = 11.84MLTSKK230 pKa = 10.56FYY232 pKa = 10.56FQSDD236 pKa = 3.69FCNVNLLGIVASACKK251 pKa = 10.34LNSPWLEE258 pKa = 3.73PDD260 pKa = 4.28KK261 pKa = 10.48ITPCITFYY269 pKa = 11.44ALKK272 pKa = 9.24NTVFKK277 pKa = 10.78NQNITEE283 pKa = 4.13AQNAYY288 pKa = 7.72TALTAIIVQNKK299 pKa = 7.48EE300 pKa = 3.39MAYY303 pKa = 8.44NTWLEE308 pKa = 4.15HH309 pKa = 5.51HH310 pKa = 6.85WGSFGNGKK318 pKa = 9.87AFNITNVKK326 pKa = 10.44DD327 pKa = 3.79NSTLTQNQINTLRR340 pKa = 11.84QTAEE344 pKa = 3.64NTFKK348 pKa = 10.91SRR350 pKa = 11.84YY351 pKa = 7.57NFIYY355 pKa = 9.86EE356 pKa = 4.2AQPPTTFTYY365 pKa = 11.11YY366 pKa = 10.88NLHH369 pKa = 5.31GWGLYY374 pKa = 10.02SPLWLDD380 pKa = 3.42PDD382 pKa = 4.21RR383 pKa = 11.84IEE385 pKa = 4.92PEE387 pKa = 3.79SEE389 pKa = 3.72AAFLAVRR396 pKa = 11.84YY397 pKa = 9.9NPLNDD402 pKa = 3.56KK403 pKa = 11.64GEE405 pKa = 4.47GNWVALQPLTKK416 pKa = 9.87EE417 pKa = 4.5LPILDD422 pKa = 4.53DD423 pKa = 3.51TCKK426 pKa = 10.0MVLKK430 pKa = 10.28DD431 pKa = 3.7YY432 pKa = 10.21PLWLILYY439 pKa = 7.08GYY441 pKa = 9.01PDD443 pKa = 4.42AATKK447 pKa = 10.12ILSGSSPLNNTRR459 pKa = 11.84LIIRR463 pKa = 11.84CQYY466 pKa = 9.6TDD468 pKa = 3.88PPLAFPQATDD478 pKa = 3.25YY479 pKa = 11.22KK480 pKa = 10.01RR481 pKa = 11.84GYY483 pKa = 8.43TIYY486 pKa = 10.83SRR488 pKa = 11.84DD489 pKa = 3.43FATGRR494 pKa = 11.84MPGGNTFIPITTARR508 pKa = 11.84LWYY511 pKa = 9.44PRR513 pKa = 11.84LQNQLDD519 pKa = 3.85VVEE522 pKa = 5.8AIVNCGPLMPRR533 pKa = 11.84DD534 pKa = 4.4DD535 pKa = 4.7LHH537 pKa = 7.64QGWAVTMGYY546 pKa = 10.29KK547 pKa = 10.22YY548 pKa = 10.47RR549 pKa = 11.84FQLGGNLPPKK559 pKa = 9.87QDD561 pKa = 3.49PVDD564 pKa = 3.64PCKK567 pKa = 10.74VPKK570 pKa = 10.29RR571 pKa = 11.84EE572 pKa = 4.22LPDD575 pKa = 3.8PSAEE579 pKa = 3.95LTAVQIVDD587 pKa = 4.16PATMDD592 pKa = 4.0PLSTLHH598 pKa = 6.26PWDD601 pKa = 3.77YY602 pKa = 10.46RR603 pKa = 11.84RR604 pKa = 11.84SMLTSSGIKK613 pKa = 10.02RR614 pKa = 11.84MSADD618 pKa = 3.81RR619 pKa = 11.84EE620 pKa = 4.43TDD622 pKa = 2.98SSLYY626 pKa = 10.26PGATGVAKK634 pKa = 10.38RR635 pKa = 11.84PKK637 pKa = 9.92TDD639 pKa = 2.72VPTQKK644 pKa = 10.73GEE646 pKa = 4.03EE647 pKa = 4.16PGPGEE652 pKa = 4.05RR653 pKa = 11.84LRR655 pKa = 11.84EE656 pKa = 3.99VLQSLLEE663 pKa = 4.07EE664 pKa = 4.28QQQEE668 pKa = 4.6TEE670 pKa = 4.97DD671 pKa = 4.32PPSPPPGEE679 pKa = 4.49RR680 pKa = 11.84VQQPEE685 pKa = 4.52LFQQLALRR693 pKa = 11.84YY694 pKa = 8.85QLQQQRR700 pKa = 11.84EE701 pKa = 4.28RR702 pKa = 11.84QGKK705 pKa = 8.09LAQGIKK711 pKa = 11.01YY712 pKa = 7.92MFQQLIKK719 pKa = 8.17TQQGVQVDD727 pKa = 3.9PRR729 pKa = 11.84LLL731 pKa = 4.26
Molecular weight: 85.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
990 |
125 |
731 |
330.0 |
37.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.061 ± 1.993 | 1.717 ± 0.833 |
4.444 ± 0.717 | 4.646 ± 0.75 |
2.424 ± 0.865 | 6.465 ± 1.645 |
1.919 ± 0.403 | 3.232 ± 1.068 |
5.152 ± 1.101 | 8.99 ± 1.525 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.919 ± 0.438 | 3.939 ± 1.034 |
7.677 ± 0.579 | 5.455 ± 1.89 |
10.202 ± 0.881 | 6.364 ± 3.995 |
8.081 ± 0.62 | 4.646 ± 1.044 |
2.828 ± 0.264 | 3.838 ± 1.275 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
