
Impatiens necrotic spot virus (INSV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Tospoviridae; Orthotospovirus; Impatiens necrotic spot orthotospovirus
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
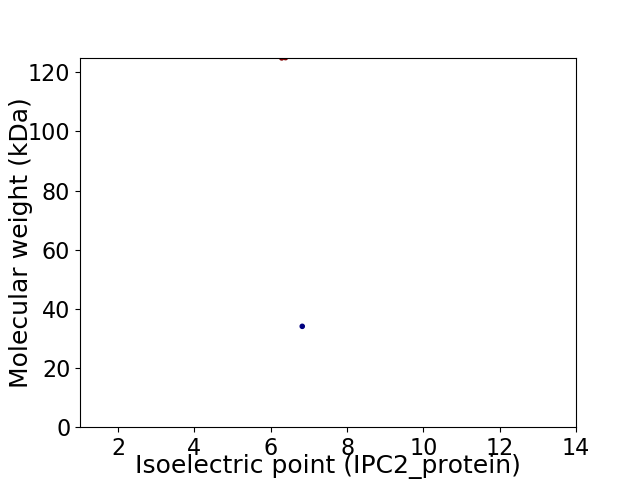
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q01268|MVP_INSV Movement protein OS=Impatiens necrotic spot virus OX=11612 PE=3 SV=1
MM1 pKa = 7.5ALKK4 pKa = 9.09EE5 pKa = 4.13TDD7 pKa = 3.12AKK9 pKa = 10.65IHH11 pKa = 5.38VEE13 pKa = 4.04RR14 pKa = 11.84GDD16 pKa = 3.39HH17 pKa = 6.65PEE19 pKa = 4.62IYY21 pKa = 10.51DD22 pKa = 3.32EE23 pKa = 4.74AYY25 pKa = 9.53YY26 pKa = 10.81DD27 pKa = 4.18RR28 pKa = 11.84SVDD31 pKa = 3.56HH32 pKa = 6.96KK33 pKa = 11.71NEE35 pKa = 3.91ILDD38 pKa = 3.82TLAEE42 pKa = 4.09MLQNATGKK50 pKa = 7.06TLRR53 pKa = 11.84PTRR56 pKa = 11.84DD57 pKa = 3.33TQTVLANNEE66 pKa = 4.16VPQSSSGLSSTPTTISIMDD85 pKa = 4.59LPNPCLNASSLTCSIKK101 pKa = 10.62GVSTFNVYY109 pKa = 10.22YY110 pKa = 9.94QVEE113 pKa = 4.75SNGVIYY119 pKa = 10.66SCISDD124 pKa = 4.49TITKK128 pKa = 10.21LGNCEE133 pKa = 4.02GSSEE137 pKa = 4.11LPRR140 pKa = 11.84SFEE143 pKa = 4.21TVPVVPITKK152 pKa = 9.22IDD154 pKa = 3.73NKK156 pKa = 10.33RR157 pKa = 11.84KK158 pKa = 10.08LSIGTKK164 pKa = 9.78FYY166 pKa = 10.47IIEE169 pKa = 4.07SLEE172 pKa = 4.07NYY174 pKa = 9.35NYY176 pKa = 10.33PIMYY180 pKa = 10.06NSRR183 pKa = 11.84PTNGTVSLQSVKK195 pKa = 10.37FSGDD199 pKa = 3.39CKK201 pKa = 10.19ISKK204 pKa = 9.05TNIVNSYY211 pKa = 8.38TVSLTTPEE219 pKa = 4.65KK220 pKa = 10.36IMGYY224 pKa = 6.84VVKK227 pKa = 10.74RR228 pKa = 11.84EE229 pKa = 3.98GSDD232 pKa = 3.15MSHH235 pKa = 7.26SIISFSGSVSLTFTEE250 pKa = 4.27EE251 pKa = 4.06NMDD254 pKa = 3.58GKK256 pKa = 10.31HH257 pKa = 6.47NLLCGDD263 pKa = 4.15KK264 pKa = 10.33SSKK267 pKa = 10.44VPLVDD272 pKa = 4.11KK273 pKa = 10.66RR274 pKa = 11.84VRR276 pKa = 11.84DD277 pKa = 4.21CIIKK281 pKa = 9.92YY282 pKa = 10.26SKK284 pKa = 10.79NIYY287 pKa = 9.4KK288 pKa = 8.29QTACINFSWFRR299 pKa = 11.84LIMIALIVYY308 pKa = 7.54FPIRR312 pKa = 11.84YY313 pKa = 8.5LVNKK317 pKa = 8.41TSKK320 pKa = 9.24TLFYY324 pKa = 10.53GYY326 pKa = 10.61DD327 pKa = 3.51LLGLITYY334 pKa = 7.77PILLLINYY342 pKa = 7.32LWSYY346 pKa = 10.77FPLKK350 pKa = 10.76CKK352 pKa = 10.73VCGNLCLVTHH362 pKa = 6.93EE363 pKa = 4.52CSKK366 pKa = 11.31LCICNKK372 pKa = 9.76NKK374 pKa = 10.29ASEE377 pKa = 4.16EE378 pKa = 3.99HH379 pKa = 7.07SEE381 pKa = 4.05EE382 pKa = 4.33CPIITRR388 pKa = 11.84TAEE391 pKa = 3.92KK392 pKa = 10.15NKK394 pKa = 9.75KK395 pKa = 8.79YY396 pKa = 10.53NWASIEE402 pKa = 4.06WFHH405 pKa = 7.61LIVNTKK411 pKa = 10.01IGLSFLKK418 pKa = 10.62AVTEE422 pKa = 4.06TLIGFLILSQMPMSMAQTAQCLDD445 pKa = 2.97SCYY448 pKa = 10.46YY449 pKa = 10.91VPGCDD454 pKa = 2.32RR455 pKa = 11.84FVTNRR460 pKa = 11.84YY461 pKa = 8.59DD462 pKa = 3.21KK463 pKa = 11.24CPEE466 pKa = 3.91KK467 pKa = 10.75DD468 pKa = 3.21QCFCAIKK475 pKa = 10.07EE476 pKa = 4.05NSIVEE481 pKa = 4.63SNFLTNVVTEE491 pKa = 4.61GPMDD495 pKa = 5.18CIPYY499 pKa = 9.07QEE501 pKa = 4.41CKK503 pKa = 10.82GRR505 pKa = 11.84ITEE508 pKa = 4.09NALVTFVKK516 pKa = 10.4CRR518 pKa = 11.84FGCEE522 pKa = 3.71YY523 pKa = 11.3ASIFQSKK530 pKa = 9.64PLDD533 pKa = 3.75NGFLEE538 pKa = 4.42YY539 pKa = 10.95SGDD542 pKa = 3.86TLGLNAVNLHH552 pKa = 4.84FMKK555 pKa = 10.75RR556 pKa = 11.84LRR558 pKa = 11.84NGIIDD563 pKa = 4.45FYY565 pKa = 11.59NKK567 pKa = 8.26TEE569 pKa = 3.88KK570 pKa = 10.57YY571 pKa = 10.71GYY573 pKa = 10.07ISGDD577 pKa = 3.16ALKK580 pKa = 10.85SNEE583 pKa = 3.86SDD585 pKa = 2.97IPEE588 pKa = 4.59SIFPRR593 pKa = 11.84KK594 pKa = 9.73SLIFDD599 pKa = 3.5SVIDD603 pKa = 3.76GKK605 pKa = 10.34YY606 pKa = 9.67RR607 pKa = 11.84YY608 pKa = 9.11MIEE611 pKa = 4.0EE612 pKa = 4.28SLLSGGGTVFSLNDD626 pKa = 3.62KK627 pKa = 10.76SSSTAQKK634 pKa = 9.78FVVYY638 pKa = 9.74IKK640 pKa = 10.38KK641 pKa = 9.93VRR643 pKa = 11.84IQYY646 pKa = 9.9DD647 pKa = 3.22VSEE650 pKa = 4.73QYY652 pKa = 7.67TTAPIQSTHH661 pKa = 5.61TDD663 pKa = 3.8FFSTCTGKK671 pKa = 10.88CSDD674 pKa = 4.38CRR676 pKa = 11.84KK677 pKa = 8.46EE678 pKa = 4.01QPITGYY684 pKa = 10.63QDD686 pKa = 3.33FCITPTSYY694 pKa = 9.62WGCEE698 pKa = 4.11EE699 pKa = 4.3VWCLAINEE707 pKa = 4.4GATCGFCRR715 pKa = 11.84NVYY718 pKa = 11.13DD719 pKa = 3.45MDD721 pKa = 3.6QSFRR725 pKa = 11.84IYY727 pKa = 10.86SVIKK731 pKa = 9.22STIKK735 pKa = 10.72SEE737 pKa = 4.06VCISGFVGAKK747 pKa = 9.44CFTVSEE753 pKa = 4.44EE754 pKa = 4.22VPSEE758 pKa = 4.0SGYY761 pKa = 10.39FQADD765 pKa = 3.04ILADD769 pKa = 3.62FHH771 pKa = 8.0NDD773 pKa = 2.74GLTIGQLIAHH783 pKa = 7.06GPDD786 pKa = 2.82SHH788 pKa = 8.61VYY790 pKa = 9.58AGNIARR796 pKa = 11.84LNNPSKK802 pKa = 10.61MFGHH806 pKa = 7.02PQLSHH811 pKa = 6.7QGDD814 pKa = 4.2PIFSKK819 pKa = 9.6KK820 pKa = 8.86TLDD823 pKa = 3.75TNDD826 pKa = 4.41LSWDD830 pKa = 3.46CSAIGKK836 pKa = 8.21KK837 pKa = 9.01TITIKK842 pKa = 10.49SCGYY846 pKa = 8.39DD847 pKa = 2.98TYY849 pKa = 11.34RR850 pKa = 11.84FKK852 pKa = 10.79TGLNQISDD860 pKa = 3.76IPVQFTDD867 pKa = 3.21QNSFYY872 pKa = 9.67MEE874 pKa = 4.96KK875 pKa = 10.06IFSLGKK881 pKa = 10.4LKK883 pKa = 10.36IVLDD887 pKa = 4.15LPSEE891 pKa = 4.15LFKK894 pKa = 10.56TVPKK898 pKa = 10.58KK899 pKa = 10.55PILSSVSLSCKK910 pKa = 10.43GCFLCSQGLRR920 pKa = 11.84CAASFISDD928 pKa = 3.16ITFSARR934 pKa = 11.84LTMKK938 pKa = 9.95QCSLSTYY945 pKa = 10.14QIAVKK950 pKa = 10.49KK951 pKa = 10.2GANKK955 pKa = 10.31YY956 pKa = 9.72NLTMFCTSNPEE967 pKa = 3.89KK968 pKa = 10.32QKK970 pKa = 10.68MIIEE974 pKa = 4.3PEE976 pKa = 3.94GDD978 pKa = 2.84KK979 pKa = 10.99SYY981 pKa = 11.38SVEE984 pKa = 3.92ALVDD988 pKa = 3.94SVAVLEE994 pKa = 4.54PEE996 pKa = 4.99NIIDD1000 pKa = 4.54QNDD1003 pKa = 3.33QHH1005 pKa = 7.97AHH1007 pKa = 6.35EE1008 pKa = 4.39EE1009 pKa = 3.97QQYY1012 pKa = 11.41NSDD1015 pKa = 3.46TSVWSFWDD1023 pKa = 3.81YY1024 pKa = 11.48VKK1026 pKa = 11.19SPFNFIASHH1035 pKa = 6.48FGSFFDD1041 pKa = 3.63TVRR1044 pKa = 11.84VVLLILFVFALAYY1057 pKa = 10.1LCSIVATMCRR1067 pKa = 11.84GYY1069 pKa = 10.84VRR1071 pKa = 11.84NKK1073 pKa = 10.21SYY1075 pKa = 8.32KK1076 pKa = 8.2TKK1078 pKa = 10.78YY1079 pKa = 10.15IEE1081 pKa = 4.29DD1082 pKa = 3.9TNDD1085 pKa = 2.96YY1086 pKa = 10.38SLVSTSSGKK1095 pKa = 8.74DD1096 pKa = 2.89TITRR1100 pKa = 11.84RR1101 pKa = 11.84RR1102 pKa = 11.84PPLDD1106 pKa = 3.3FSGII1110 pKa = 3.76
MM1 pKa = 7.5ALKK4 pKa = 9.09EE5 pKa = 4.13TDD7 pKa = 3.12AKK9 pKa = 10.65IHH11 pKa = 5.38VEE13 pKa = 4.04RR14 pKa = 11.84GDD16 pKa = 3.39HH17 pKa = 6.65PEE19 pKa = 4.62IYY21 pKa = 10.51DD22 pKa = 3.32EE23 pKa = 4.74AYY25 pKa = 9.53YY26 pKa = 10.81DD27 pKa = 4.18RR28 pKa = 11.84SVDD31 pKa = 3.56HH32 pKa = 6.96KK33 pKa = 11.71NEE35 pKa = 3.91ILDD38 pKa = 3.82TLAEE42 pKa = 4.09MLQNATGKK50 pKa = 7.06TLRR53 pKa = 11.84PTRR56 pKa = 11.84DD57 pKa = 3.33TQTVLANNEE66 pKa = 4.16VPQSSSGLSSTPTTISIMDD85 pKa = 4.59LPNPCLNASSLTCSIKK101 pKa = 10.62GVSTFNVYY109 pKa = 10.22YY110 pKa = 9.94QVEE113 pKa = 4.75SNGVIYY119 pKa = 10.66SCISDD124 pKa = 4.49TITKK128 pKa = 10.21LGNCEE133 pKa = 4.02GSSEE137 pKa = 4.11LPRR140 pKa = 11.84SFEE143 pKa = 4.21TVPVVPITKK152 pKa = 9.22IDD154 pKa = 3.73NKK156 pKa = 10.33RR157 pKa = 11.84KK158 pKa = 10.08LSIGTKK164 pKa = 9.78FYY166 pKa = 10.47IIEE169 pKa = 4.07SLEE172 pKa = 4.07NYY174 pKa = 9.35NYY176 pKa = 10.33PIMYY180 pKa = 10.06NSRR183 pKa = 11.84PTNGTVSLQSVKK195 pKa = 10.37FSGDD199 pKa = 3.39CKK201 pKa = 10.19ISKK204 pKa = 9.05TNIVNSYY211 pKa = 8.38TVSLTTPEE219 pKa = 4.65KK220 pKa = 10.36IMGYY224 pKa = 6.84VVKK227 pKa = 10.74RR228 pKa = 11.84EE229 pKa = 3.98GSDD232 pKa = 3.15MSHH235 pKa = 7.26SIISFSGSVSLTFTEE250 pKa = 4.27EE251 pKa = 4.06NMDD254 pKa = 3.58GKK256 pKa = 10.31HH257 pKa = 6.47NLLCGDD263 pKa = 4.15KK264 pKa = 10.33SSKK267 pKa = 10.44VPLVDD272 pKa = 4.11KK273 pKa = 10.66RR274 pKa = 11.84VRR276 pKa = 11.84DD277 pKa = 4.21CIIKK281 pKa = 9.92YY282 pKa = 10.26SKK284 pKa = 10.79NIYY287 pKa = 9.4KK288 pKa = 8.29QTACINFSWFRR299 pKa = 11.84LIMIALIVYY308 pKa = 7.54FPIRR312 pKa = 11.84YY313 pKa = 8.5LVNKK317 pKa = 8.41TSKK320 pKa = 9.24TLFYY324 pKa = 10.53GYY326 pKa = 10.61DD327 pKa = 3.51LLGLITYY334 pKa = 7.77PILLLINYY342 pKa = 7.32LWSYY346 pKa = 10.77FPLKK350 pKa = 10.76CKK352 pKa = 10.73VCGNLCLVTHH362 pKa = 6.93EE363 pKa = 4.52CSKK366 pKa = 11.31LCICNKK372 pKa = 9.76NKK374 pKa = 10.29ASEE377 pKa = 4.16EE378 pKa = 3.99HH379 pKa = 7.07SEE381 pKa = 4.05EE382 pKa = 4.33CPIITRR388 pKa = 11.84TAEE391 pKa = 3.92KK392 pKa = 10.15NKK394 pKa = 9.75KK395 pKa = 8.79YY396 pKa = 10.53NWASIEE402 pKa = 4.06WFHH405 pKa = 7.61LIVNTKK411 pKa = 10.01IGLSFLKK418 pKa = 10.62AVTEE422 pKa = 4.06TLIGFLILSQMPMSMAQTAQCLDD445 pKa = 2.97SCYY448 pKa = 10.46YY449 pKa = 10.91VPGCDD454 pKa = 2.32RR455 pKa = 11.84FVTNRR460 pKa = 11.84YY461 pKa = 8.59DD462 pKa = 3.21KK463 pKa = 11.24CPEE466 pKa = 3.91KK467 pKa = 10.75DD468 pKa = 3.21QCFCAIKK475 pKa = 10.07EE476 pKa = 4.05NSIVEE481 pKa = 4.63SNFLTNVVTEE491 pKa = 4.61GPMDD495 pKa = 5.18CIPYY499 pKa = 9.07QEE501 pKa = 4.41CKK503 pKa = 10.82GRR505 pKa = 11.84ITEE508 pKa = 4.09NALVTFVKK516 pKa = 10.4CRR518 pKa = 11.84FGCEE522 pKa = 3.71YY523 pKa = 11.3ASIFQSKK530 pKa = 9.64PLDD533 pKa = 3.75NGFLEE538 pKa = 4.42YY539 pKa = 10.95SGDD542 pKa = 3.86TLGLNAVNLHH552 pKa = 4.84FMKK555 pKa = 10.75RR556 pKa = 11.84LRR558 pKa = 11.84NGIIDD563 pKa = 4.45FYY565 pKa = 11.59NKK567 pKa = 8.26TEE569 pKa = 3.88KK570 pKa = 10.57YY571 pKa = 10.71GYY573 pKa = 10.07ISGDD577 pKa = 3.16ALKK580 pKa = 10.85SNEE583 pKa = 3.86SDD585 pKa = 2.97IPEE588 pKa = 4.59SIFPRR593 pKa = 11.84KK594 pKa = 9.73SLIFDD599 pKa = 3.5SVIDD603 pKa = 3.76GKK605 pKa = 10.34YY606 pKa = 9.67RR607 pKa = 11.84YY608 pKa = 9.11MIEE611 pKa = 4.0EE612 pKa = 4.28SLLSGGGTVFSLNDD626 pKa = 3.62KK627 pKa = 10.76SSSTAQKK634 pKa = 9.78FVVYY638 pKa = 9.74IKK640 pKa = 10.38KK641 pKa = 9.93VRR643 pKa = 11.84IQYY646 pKa = 9.9DD647 pKa = 3.22VSEE650 pKa = 4.73QYY652 pKa = 7.67TTAPIQSTHH661 pKa = 5.61TDD663 pKa = 3.8FFSTCTGKK671 pKa = 10.88CSDD674 pKa = 4.38CRR676 pKa = 11.84KK677 pKa = 8.46EE678 pKa = 4.01QPITGYY684 pKa = 10.63QDD686 pKa = 3.33FCITPTSYY694 pKa = 9.62WGCEE698 pKa = 4.11EE699 pKa = 4.3VWCLAINEE707 pKa = 4.4GATCGFCRR715 pKa = 11.84NVYY718 pKa = 11.13DD719 pKa = 3.45MDD721 pKa = 3.6QSFRR725 pKa = 11.84IYY727 pKa = 10.86SVIKK731 pKa = 9.22STIKK735 pKa = 10.72SEE737 pKa = 4.06VCISGFVGAKK747 pKa = 9.44CFTVSEE753 pKa = 4.44EE754 pKa = 4.22VPSEE758 pKa = 4.0SGYY761 pKa = 10.39FQADD765 pKa = 3.04ILADD769 pKa = 3.62FHH771 pKa = 8.0NDD773 pKa = 2.74GLTIGQLIAHH783 pKa = 7.06GPDD786 pKa = 2.82SHH788 pKa = 8.61VYY790 pKa = 9.58AGNIARR796 pKa = 11.84LNNPSKK802 pKa = 10.61MFGHH806 pKa = 7.02PQLSHH811 pKa = 6.7QGDD814 pKa = 4.2PIFSKK819 pKa = 9.6KK820 pKa = 8.86TLDD823 pKa = 3.75TNDD826 pKa = 4.41LSWDD830 pKa = 3.46CSAIGKK836 pKa = 8.21KK837 pKa = 9.01TITIKK842 pKa = 10.49SCGYY846 pKa = 8.39DD847 pKa = 2.98TYY849 pKa = 11.34RR850 pKa = 11.84FKK852 pKa = 10.79TGLNQISDD860 pKa = 3.76IPVQFTDD867 pKa = 3.21QNSFYY872 pKa = 9.67MEE874 pKa = 4.96KK875 pKa = 10.06IFSLGKK881 pKa = 10.4LKK883 pKa = 10.36IVLDD887 pKa = 4.15LPSEE891 pKa = 4.15LFKK894 pKa = 10.56TVPKK898 pKa = 10.58KK899 pKa = 10.55PILSSVSLSCKK910 pKa = 10.43GCFLCSQGLRR920 pKa = 11.84CAASFISDD928 pKa = 3.16ITFSARR934 pKa = 11.84LTMKK938 pKa = 9.95QCSLSTYY945 pKa = 10.14QIAVKK950 pKa = 10.49KK951 pKa = 10.2GANKK955 pKa = 10.31YY956 pKa = 9.72NLTMFCTSNPEE967 pKa = 3.89KK968 pKa = 10.32QKK970 pKa = 10.68MIIEE974 pKa = 4.3PEE976 pKa = 3.94GDD978 pKa = 2.84KK979 pKa = 10.99SYY981 pKa = 11.38SVEE984 pKa = 3.92ALVDD988 pKa = 3.94SVAVLEE994 pKa = 4.54PEE996 pKa = 4.99NIIDD1000 pKa = 4.54QNDD1003 pKa = 3.33QHH1005 pKa = 7.97AHH1007 pKa = 6.35EE1008 pKa = 4.39EE1009 pKa = 3.97QQYY1012 pKa = 11.41NSDD1015 pKa = 3.46TSVWSFWDD1023 pKa = 3.81YY1024 pKa = 11.48VKK1026 pKa = 11.19SPFNFIASHH1035 pKa = 6.48FGSFFDD1041 pKa = 3.63TVRR1044 pKa = 11.84VVLLILFVFALAYY1057 pKa = 10.1LCSIVATMCRR1067 pKa = 11.84GYY1069 pKa = 10.84VRR1071 pKa = 11.84NKK1073 pKa = 10.21SYY1075 pKa = 8.32KK1076 pKa = 8.2TKK1078 pKa = 10.78YY1079 pKa = 10.15IEE1081 pKa = 4.29DD1082 pKa = 3.9TNDD1085 pKa = 2.96YY1086 pKa = 10.38SLVSTSSGKK1095 pKa = 8.74DD1096 pKa = 2.89TITRR1100 pKa = 11.84RR1101 pKa = 11.84RR1102 pKa = 11.84PPLDD1106 pKa = 3.3FSGII1110 pKa = 3.76
Molecular weight: 124.87 kDa
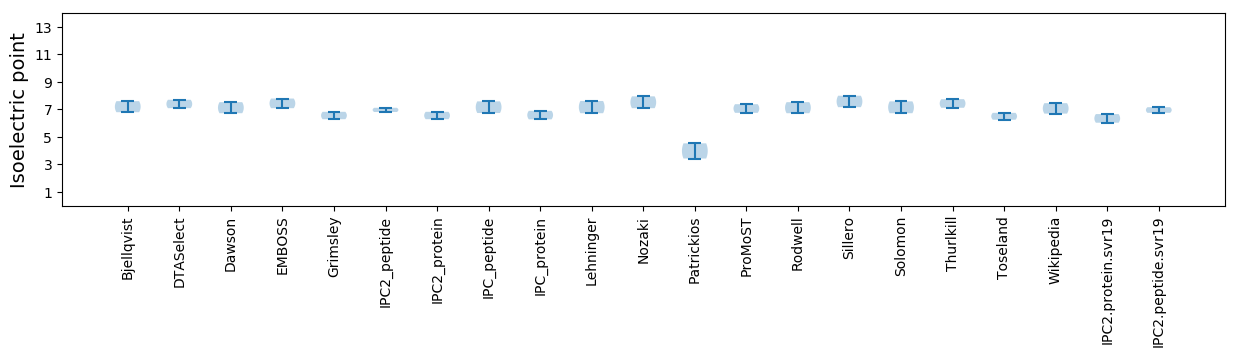
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q01268|MVP_INSV Movement protein OS=Impatiens necrotic spot virus OX=11612 PE=3 SV=1
MM1 pKa = 7.48NSFFKK6 pKa = 10.63SLRR9 pKa = 11.84SSSSRR14 pKa = 11.84EE15 pKa = 3.47LDD17 pKa = 3.28HH18 pKa = 7.32PRR20 pKa = 11.84VTTTLSKK27 pKa = 10.5QGADD31 pKa = 2.45IVVHH35 pKa = 6.1NPSANHH41 pKa = 5.76NNKK44 pKa = 9.51EE45 pKa = 4.1VLQRR49 pKa = 11.84AMDD52 pKa = 3.77SSKK55 pKa = 11.32GKK57 pKa = 10.18ILMNNTGTSSLGTYY71 pKa = 10.02EE72 pKa = 4.11SDD74 pKa = 3.4QISEE78 pKa = 4.31SEE80 pKa = 4.44SYY82 pKa = 10.89DD83 pKa = 3.18LSARR87 pKa = 11.84MIVDD91 pKa = 3.93TNHH94 pKa = 7.28HH95 pKa = 5.74ISSWKK100 pKa = 9.82NDD102 pKa = 3.42LFVGNGDD109 pKa = 3.39KK110 pKa = 10.99AATKK114 pKa = 9.89IIKK117 pKa = 9.32IHH119 pKa = 5.57PTWDD123 pKa = 3.06SRR125 pKa = 11.84KK126 pKa = 8.54QYY128 pKa = 10.8MMISRR133 pKa = 11.84IVIWICPTIADD144 pKa = 4.47PDD146 pKa = 3.85GKK148 pKa = 10.69LAVALIDD155 pKa = 3.91PNKK158 pKa = 10.21SVNARR163 pKa = 11.84TVLKK167 pKa = 10.4GQGSIKK173 pKa = 10.74DD174 pKa = 3.77PICFVFYY181 pKa = 10.61LNWSIPKK188 pKa = 9.92VNNTSEE194 pKa = 4.29NCVQLHH200 pKa = 6.15LLCDD204 pKa = 3.46QVYY207 pKa = 10.33KK208 pKa = 10.68KK209 pKa = 10.4DD210 pKa = 3.56VSFASVMYY218 pKa = 9.84SWTKK222 pKa = 9.77EE223 pKa = 3.95FCDD226 pKa = 4.72SPRR229 pKa = 11.84ADD231 pKa = 4.0LDD233 pKa = 3.82KK234 pKa = 11.18SCMIIPINRR243 pKa = 11.84AIRR246 pKa = 11.84AKK248 pKa = 9.89SQAFIEE254 pKa = 4.45ACKK257 pKa = 10.62LIIPKK262 pKa = 10.35GNSEE266 pKa = 3.96KK267 pKa = 10.61QIRR270 pKa = 11.84RR271 pKa = 11.84QLAEE275 pKa = 3.85LSANLEE281 pKa = 3.98KK282 pKa = 10.85SVEE285 pKa = 4.1EE286 pKa = 4.31EE287 pKa = 4.85EE288 pKa = 4.85NVTDD292 pKa = 3.94NKK294 pKa = 10.67IEE296 pKa = 3.8ISFDD300 pKa = 3.43NEE302 pKa = 3.48II303 pKa = 4.09
MM1 pKa = 7.48NSFFKK6 pKa = 10.63SLRR9 pKa = 11.84SSSSRR14 pKa = 11.84EE15 pKa = 3.47LDD17 pKa = 3.28HH18 pKa = 7.32PRR20 pKa = 11.84VTTTLSKK27 pKa = 10.5QGADD31 pKa = 2.45IVVHH35 pKa = 6.1NPSANHH41 pKa = 5.76NNKK44 pKa = 9.51EE45 pKa = 4.1VLQRR49 pKa = 11.84AMDD52 pKa = 3.77SSKK55 pKa = 11.32GKK57 pKa = 10.18ILMNNTGTSSLGTYY71 pKa = 10.02EE72 pKa = 4.11SDD74 pKa = 3.4QISEE78 pKa = 4.31SEE80 pKa = 4.44SYY82 pKa = 10.89DD83 pKa = 3.18LSARR87 pKa = 11.84MIVDD91 pKa = 3.93TNHH94 pKa = 7.28HH95 pKa = 5.74ISSWKK100 pKa = 9.82NDD102 pKa = 3.42LFVGNGDD109 pKa = 3.39KK110 pKa = 10.99AATKK114 pKa = 9.89IIKK117 pKa = 9.32IHH119 pKa = 5.57PTWDD123 pKa = 3.06SRR125 pKa = 11.84KK126 pKa = 8.54QYY128 pKa = 10.8MMISRR133 pKa = 11.84IVIWICPTIADD144 pKa = 4.47PDD146 pKa = 3.85GKK148 pKa = 10.69LAVALIDD155 pKa = 3.91PNKK158 pKa = 10.21SVNARR163 pKa = 11.84TVLKK167 pKa = 10.4GQGSIKK173 pKa = 10.74DD174 pKa = 3.77PICFVFYY181 pKa = 10.61LNWSIPKK188 pKa = 9.92VNNTSEE194 pKa = 4.29NCVQLHH200 pKa = 6.15LLCDD204 pKa = 3.46QVYY207 pKa = 10.33KK208 pKa = 10.68KK209 pKa = 10.4DD210 pKa = 3.56VSFASVMYY218 pKa = 9.84SWTKK222 pKa = 9.77EE223 pKa = 3.95FCDD226 pKa = 4.72SPRR229 pKa = 11.84ADD231 pKa = 4.0LDD233 pKa = 3.82KK234 pKa = 11.18SCMIIPINRR243 pKa = 11.84AIRR246 pKa = 11.84AKK248 pKa = 9.89SQAFIEE254 pKa = 4.45ACKK257 pKa = 10.62LIIPKK262 pKa = 10.35GNSEE266 pKa = 3.96KK267 pKa = 10.61QIRR270 pKa = 11.84RR271 pKa = 11.84QLAEE275 pKa = 3.85LSANLEE281 pKa = 3.98KK282 pKa = 10.85SVEE285 pKa = 4.1EE286 pKa = 4.31EE287 pKa = 4.85EE288 pKa = 4.85NVTDD292 pKa = 3.94NKK294 pKa = 10.67IEE296 pKa = 3.8ISFDD300 pKa = 3.43NEE302 pKa = 3.48II303 pKa = 4.09
Molecular weight: 34.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1413 |
303 |
1110 |
706.5 |
79.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.317 ± 0.808 | 3.68 ± 0.682 |
5.662 ± 0.468 | 5.308 ± 0.151 |
4.6 ± 0.812 | 4.954 ± 0.823 |
1.769 ± 0.269 | 8.209 ± 0.514 |
7.502 ± 0.373 | 7.431 ± 0.413 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.052 ± 0.293 | 5.662 ± 0.796 |
3.822 ± 0.095 | 3.114 ± 0.093 |
3.397 ± 0.445 | 10.616 ± 0.466 |
6.582 ± 0.977 | 6.016 ± 0.037 |
0.991 ± 0.328 | 4.317 ± 1.164 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
