
Red clover powdery mildew-associated totivirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
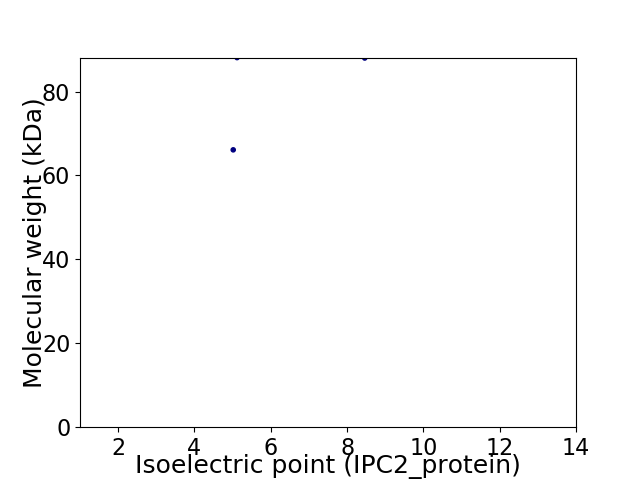
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
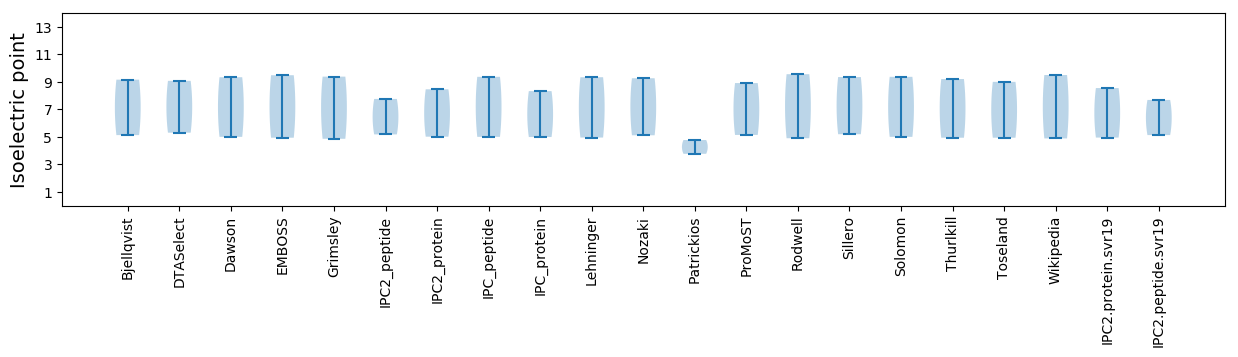
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S3Q2X1|A0A0S3Q2X1_9VIRU RNA-directed RNA polymerase OS=Red clover powdery mildew-associated totivirus 2 OX=1714363 GN=RdRp PE=3 SV=1
MM1 pKa = 8.05DD2 pKa = 4.69SMVFDD7 pKa = 3.69TCVDD11 pKa = 3.43NVTSLLYY18 pKa = 10.58NLLRR22 pKa = 11.84MYY24 pKa = 10.74YY25 pKa = 9.71ILEE28 pKa = 4.26NKK30 pKa = 9.17NLSVKK35 pKa = 9.49KK36 pKa = 9.0TDD38 pKa = 2.91SYY40 pKa = 11.77YY41 pKa = 10.92DD42 pKa = 4.14DD43 pKa = 3.82GHH45 pKa = 8.39LSVDD49 pKa = 3.17MGDD52 pKa = 3.22VFGVGEE58 pKa = 4.03RR59 pKa = 11.84SKK61 pKa = 10.97RR62 pKa = 11.84ITDD65 pKa = 3.28VRR67 pKa = 11.84FNPCRR72 pKa = 11.84AHH74 pKa = 7.34RR75 pKa = 11.84YY76 pKa = 8.76DD77 pKa = 3.84AEE79 pKa = 4.64TYY81 pKa = 9.53FLNFSGEE88 pKa = 4.43PVLHH92 pKa = 6.15MANANALSSGILATALGEE110 pKa = 4.28WSSEE114 pKa = 3.75TPYY117 pKa = 10.75AISHH121 pKa = 7.01PSPALAQNIIINTRR135 pKa = 11.84GGVSQDD141 pKa = 3.46EE142 pKa = 4.3IEE144 pKa = 4.58SFNQITTNDD153 pKa = 3.44VLKK156 pKa = 10.54VIKK159 pKa = 10.44DD160 pKa = 3.31IVEE163 pKa = 4.19SNRR166 pKa = 11.84YY167 pKa = 8.02HH168 pKa = 6.47AQFEE172 pKa = 4.34LAYY175 pKa = 11.22LMLAQLLFTLKK186 pKa = 10.12PRR188 pKa = 11.84SAEE191 pKa = 3.74ALIWMHH197 pKa = 6.01GTPLMQLPKK206 pKa = 10.73FMTFRR211 pKa = 11.84GLVNEE216 pKa = 4.49TVSLAPYY223 pKa = 9.95VPFPEE228 pKa = 5.26RR229 pKa = 11.84LDD231 pKa = 3.82TYY233 pKa = 8.94GTWLANKK240 pKa = 9.0EE241 pKa = 3.99AIYY244 pKa = 10.4YY245 pKa = 10.22HH246 pKa = 6.49SLLINEE252 pKa = 4.52VVMTYY257 pKa = 10.3AYY259 pKa = 9.97EE260 pKa = 5.13AITTNDD266 pKa = 2.65ITRR269 pKa = 11.84LLLRR273 pKa = 11.84QIGGIFSAEE282 pKa = 4.07PGLMADD288 pKa = 4.29MSCFAQVLQKK298 pKa = 10.75EE299 pKa = 4.44VLLPFEE305 pKa = 4.69TNGGLDD311 pKa = 3.0RR312 pKa = 11.84FTWINCLDD320 pKa = 4.05GEE322 pKa = 4.35IGMQVKK328 pKa = 10.41VNDD331 pKa = 4.23PNAQHH336 pKa = 7.22HH337 pKa = 6.09YY338 pKa = 10.68RR339 pKa = 11.84LDD341 pKa = 3.65TLEE344 pKa = 5.56LDD346 pKa = 4.32DD347 pKa = 4.65GVIRR351 pKa = 11.84HH352 pKa = 5.79TLRR355 pKa = 11.84LDD357 pKa = 3.29SFVPYY362 pKa = 10.09VYY364 pKa = 10.15PVLLYY369 pKa = 10.48GYY371 pKa = 10.07APSTYY376 pKa = 9.32YY377 pKa = 10.96QNDD380 pKa = 3.69CVRR383 pKa = 11.84EE384 pKa = 3.96VEE386 pKa = 4.29MKK388 pKa = 10.05IDD390 pKa = 3.7SKK392 pKa = 10.48KK393 pKa = 10.55EE394 pKa = 3.7RR395 pKa = 11.84MITEE399 pKa = 4.21SVDD402 pKa = 3.86DD403 pKa = 4.2FSKK406 pKa = 11.27VMNILRR412 pKa = 11.84MAGYY416 pKa = 9.64DD417 pKa = 3.41ATGGSCYY424 pKa = 10.57EE425 pKa = 4.02GDD427 pKa = 4.58KK428 pKa = 10.51VKK430 pKa = 10.83NWADD434 pKa = 3.15NSSGHH439 pKa = 5.97FLYY442 pKa = 10.33TRR444 pKa = 11.84SAEE447 pKa = 4.22APDD450 pKa = 5.03DD451 pKa = 4.76DD452 pKa = 4.97IFYY455 pKa = 10.99VDD457 pKa = 4.68LGLEE461 pKa = 4.22EE462 pKa = 5.04RR463 pKa = 11.84SKK465 pKa = 11.08SWVDD469 pKa = 3.47EE470 pKa = 4.22PRR472 pKa = 11.84WSEE475 pKa = 4.1KK476 pKa = 10.34ACLRR480 pKa = 11.84SEE482 pKa = 4.35VKK484 pKa = 9.04ITAFTMFKK492 pKa = 10.62DD493 pKa = 3.26GVMYY497 pKa = 10.9KK498 pKa = 10.63NITGVLKK505 pKa = 10.43KK506 pKa = 10.83YY507 pKa = 9.97SFKK510 pKa = 10.88QPSLEE515 pKa = 3.82ITSKK519 pKa = 8.75HH520 pKa = 3.92TRR522 pKa = 11.84EE523 pKa = 4.19VKK525 pKa = 9.11VTLRR529 pKa = 11.84PRR531 pKa = 11.84PHH533 pKa = 6.43IKK535 pKa = 10.29AGFRR539 pKa = 11.84MARR542 pKa = 11.84IDD544 pKa = 4.51DD545 pKa = 3.85RR546 pKa = 11.84TILHH550 pKa = 6.83PVTNTLFGDD559 pKa = 4.45LAQSDD564 pKa = 4.66EE565 pKa = 4.03QLDD568 pKa = 3.96WYY570 pKa = 10.65DD571 pKa = 3.4QDD573 pKa = 4.83QEE575 pKa = 4.78DD576 pKa = 4.24GTGAA580 pKa = 3.47
MM1 pKa = 8.05DD2 pKa = 4.69SMVFDD7 pKa = 3.69TCVDD11 pKa = 3.43NVTSLLYY18 pKa = 10.58NLLRR22 pKa = 11.84MYY24 pKa = 10.74YY25 pKa = 9.71ILEE28 pKa = 4.26NKK30 pKa = 9.17NLSVKK35 pKa = 9.49KK36 pKa = 9.0TDD38 pKa = 2.91SYY40 pKa = 11.77YY41 pKa = 10.92DD42 pKa = 4.14DD43 pKa = 3.82GHH45 pKa = 8.39LSVDD49 pKa = 3.17MGDD52 pKa = 3.22VFGVGEE58 pKa = 4.03RR59 pKa = 11.84SKK61 pKa = 10.97RR62 pKa = 11.84ITDD65 pKa = 3.28VRR67 pKa = 11.84FNPCRR72 pKa = 11.84AHH74 pKa = 7.34RR75 pKa = 11.84YY76 pKa = 8.76DD77 pKa = 3.84AEE79 pKa = 4.64TYY81 pKa = 9.53FLNFSGEE88 pKa = 4.43PVLHH92 pKa = 6.15MANANALSSGILATALGEE110 pKa = 4.28WSSEE114 pKa = 3.75TPYY117 pKa = 10.75AISHH121 pKa = 7.01PSPALAQNIIINTRR135 pKa = 11.84GGVSQDD141 pKa = 3.46EE142 pKa = 4.3IEE144 pKa = 4.58SFNQITTNDD153 pKa = 3.44VLKK156 pKa = 10.54VIKK159 pKa = 10.44DD160 pKa = 3.31IVEE163 pKa = 4.19SNRR166 pKa = 11.84YY167 pKa = 8.02HH168 pKa = 6.47AQFEE172 pKa = 4.34LAYY175 pKa = 11.22LMLAQLLFTLKK186 pKa = 10.12PRR188 pKa = 11.84SAEE191 pKa = 3.74ALIWMHH197 pKa = 6.01GTPLMQLPKK206 pKa = 10.73FMTFRR211 pKa = 11.84GLVNEE216 pKa = 4.49TVSLAPYY223 pKa = 9.95VPFPEE228 pKa = 5.26RR229 pKa = 11.84LDD231 pKa = 3.82TYY233 pKa = 8.94GTWLANKK240 pKa = 9.0EE241 pKa = 3.99AIYY244 pKa = 10.4YY245 pKa = 10.22HH246 pKa = 6.49SLLINEE252 pKa = 4.52VVMTYY257 pKa = 10.3AYY259 pKa = 9.97EE260 pKa = 5.13AITTNDD266 pKa = 2.65ITRR269 pKa = 11.84LLLRR273 pKa = 11.84QIGGIFSAEE282 pKa = 4.07PGLMADD288 pKa = 4.29MSCFAQVLQKK298 pKa = 10.75EE299 pKa = 4.44VLLPFEE305 pKa = 4.69TNGGLDD311 pKa = 3.0RR312 pKa = 11.84FTWINCLDD320 pKa = 4.05GEE322 pKa = 4.35IGMQVKK328 pKa = 10.41VNDD331 pKa = 4.23PNAQHH336 pKa = 7.22HH337 pKa = 6.09YY338 pKa = 10.68RR339 pKa = 11.84LDD341 pKa = 3.65TLEE344 pKa = 5.56LDD346 pKa = 4.32DD347 pKa = 4.65GVIRR351 pKa = 11.84HH352 pKa = 5.79TLRR355 pKa = 11.84LDD357 pKa = 3.29SFVPYY362 pKa = 10.09VYY364 pKa = 10.15PVLLYY369 pKa = 10.48GYY371 pKa = 10.07APSTYY376 pKa = 9.32YY377 pKa = 10.96QNDD380 pKa = 3.69CVRR383 pKa = 11.84EE384 pKa = 3.96VEE386 pKa = 4.29MKK388 pKa = 10.05IDD390 pKa = 3.7SKK392 pKa = 10.48KK393 pKa = 10.55EE394 pKa = 3.7RR395 pKa = 11.84MITEE399 pKa = 4.21SVDD402 pKa = 3.86DD403 pKa = 4.2FSKK406 pKa = 11.27VMNILRR412 pKa = 11.84MAGYY416 pKa = 9.64DD417 pKa = 3.41ATGGSCYY424 pKa = 10.57EE425 pKa = 4.02GDD427 pKa = 4.58KK428 pKa = 10.51VKK430 pKa = 10.83NWADD434 pKa = 3.15NSSGHH439 pKa = 5.97FLYY442 pKa = 10.33TRR444 pKa = 11.84SAEE447 pKa = 4.22APDD450 pKa = 5.03DD451 pKa = 4.76DD452 pKa = 4.97IFYY455 pKa = 10.99VDD457 pKa = 4.68LGLEE461 pKa = 4.22EE462 pKa = 5.04RR463 pKa = 11.84SKK465 pKa = 11.08SWVDD469 pKa = 3.47EE470 pKa = 4.22PRR472 pKa = 11.84WSEE475 pKa = 4.1KK476 pKa = 10.34ACLRR480 pKa = 11.84SEE482 pKa = 4.35VKK484 pKa = 9.04ITAFTMFKK492 pKa = 10.62DD493 pKa = 3.26GVMYY497 pKa = 10.9KK498 pKa = 10.63NITGVLKK505 pKa = 10.43KK506 pKa = 10.83YY507 pKa = 9.97SFKK510 pKa = 10.88QPSLEE515 pKa = 3.82ITSKK519 pKa = 8.75HH520 pKa = 3.92TRR522 pKa = 11.84EE523 pKa = 4.19VKK525 pKa = 9.11VTLRR529 pKa = 11.84PRR531 pKa = 11.84PHH533 pKa = 6.43IKK535 pKa = 10.29AGFRR539 pKa = 11.84MARR542 pKa = 11.84IDD544 pKa = 4.51DD545 pKa = 3.85RR546 pKa = 11.84TILHH550 pKa = 6.83PVTNTLFGDD559 pKa = 4.45LAQSDD564 pKa = 4.66EE565 pKa = 4.03QLDD568 pKa = 3.96WYY570 pKa = 10.65DD571 pKa = 3.4QDD573 pKa = 4.83QEE575 pKa = 4.78DD576 pKa = 4.24GTGAA580 pKa = 3.47
Molecular weight: 66.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S3Q2X1|A0A0S3Q2X1_9VIRU RNA-directed RNA polymerase OS=Red clover powdery mildew-associated totivirus 2 OX=1714363 GN=RdRp PE=3 SV=1
MM1 pKa = 7.82LITVYY6 pKa = 10.77ILMGLRR12 pKa = 11.84TRR14 pKa = 11.84GAVDD18 pKa = 3.26EE19 pKa = 4.79ALLNVKK25 pKa = 9.57ALHH28 pKa = 6.45KK29 pKa = 10.8LHH31 pKa = 6.07GQYY34 pKa = 11.01VLAIPVLFGNVIAYY48 pKa = 9.61YY49 pKa = 10.23VDD51 pKa = 3.15KK52 pKa = 10.99HH53 pKa = 6.43DD54 pKa = 3.84VTMADD59 pKa = 3.36KK60 pKa = 10.74KK61 pKa = 9.99VAKK64 pKa = 9.89QAGLVIANNVFVDD77 pKa = 4.74FCPDD81 pKa = 2.84ARR83 pKa = 11.84VDD85 pKa = 3.84YY86 pKa = 10.22IAVMGNMAKK95 pKa = 10.36GLHH98 pKa = 6.04KK99 pKa = 9.66KK100 pKa = 7.74TRR102 pKa = 11.84INDD105 pKa = 3.53RR106 pKa = 11.84EE107 pKa = 4.37LIRR110 pKa = 11.84SRR112 pKa = 11.84ISQRR116 pKa = 11.84HH117 pKa = 4.8HH118 pKa = 4.37MHH120 pKa = 6.46IKK122 pKa = 10.42VGMLDD127 pKa = 3.57SEE129 pKa = 4.33GDD131 pKa = 3.59EE132 pKa = 4.96NEE134 pKa = 4.16IEE136 pKa = 4.01LDD138 pKa = 3.75GVKK141 pKa = 10.43GYY143 pKa = 10.65IMHH146 pKa = 7.71RR147 pKa = 11.84LMKK150 pKa = 8.33MTTVRR155 pKa = 11.84QTVTTFLLYY164 pKa = 10.77VEE166 pKa = 5.6LAPSWAADD174 pKa = 3.41VLAVIVSRR182 pKa = 11.84HH183 pKa = 5.1SEE185 pKa = 3.67WAQLMDD191 pKa = 4.26DD192 pKa = 4.49LKK194 pKa = 11.09LFSMYY199 pKa = 10.01VKK201 pKa = 10.35QYY203 pKa = 8.45QHH205 pKa = 6.85VIRR208 pKa = 11.84SDD210 pKa = 3.53LAVLFEE216 pKa = 5.2LNVLANRR223 pKa = 11.84SDD225 pKa = 4.94SEE227 pKa = 4.22MDD229 pKa = 2.96WAADD233 pKa = 3.0IAKK236 pKa = 8.28RR237 pKa = 11.84TKK239 pKa = 10.68SVVATPDD246 pKa = 3.07QEE248 pKa = 4.24RR249 pKa = 11.84LYY251 pKa = 10.8RR252 pKa = 11.84DD253 pKa = 3.66GVSMFTSARR262 pKa = 11.84AEE264 pKa = 4.22GKK266 pKa = 10.13QPVTMKK272 pKa = 9.93WEE274 pKa = 4.57EE275 pKa = 4.0YY276 pKa = 10.98RR277 pKa = 11.84NMTWGLTPPGAVHH290 pKa = 6.61SPEE293 pKa = 3.71GDD295 pKa = 3.14FRR297 pKa = 11.84RR298 pKa = 11.84NISTIPSKK306 pKa = 10.83LRR308 pKa = 11.84NKK310 pKa = 8.61KK311 pKa = 8.27TYY313 pKa = 10.79NSVFRR318 pKa = 11.84PKK320 pKa = 10.26EE321 pKa = 3.79LKK323 pKa = 10.2HH324 pKa = 7.45YY325 pKa = 7.33MAQRR329 pKa = 11.84PKK331 pKa = 10.3IIAKK335 pKa = 10.06PSVKK339 pKa = 10.66YY340 pKa = 9.97EE341 pKa = 3.7WGKK344 pKa = 9.93NRR346 pKa = 11.84ALYY349 pKa = 10.66GCDD352 pKa = 3.4LVSHH356 pKa = 6.54MMTDD360 pKa = 3.87FGMMQAEE367 pKa = 4.39STLPSWMPTGRR378 pKa = 11.84MADD381 pKa = 3.48EE382 pKa = 4.27NRR384 pKa = 11.84VKK386 pKa = 10.68RR387 pKa = 11.84QIATMGNGIPFCYY400 pKa = 10.51DD401 pKa = 2.8FDD403 pKa = 4.94DD404 pKa = 4.89FNSQHH409 pKa = 5.81SVKK412 pKa = 10.75SMQTVITAWYY422 pKa = 9.13SVYY425 pKa = 11.08GDD427 pKa = 5.26LISDD431 pKa = 3.84EE432 pKa = 4.44QKK434 pKa = 11.15LCVDD438 pKa = 4.04WVVQSLEE445 pKa = 3.92TQLIINSVGLGSSKK459 pKa = 10.72YY460 pKa = 9.92IEE462 pKa = 3.96VDD464 pKa = 3.04GTLFSGWRR472 pKa = 11.84LTSFMNTCLNYY483 pKa = 10.7LYY485 pKa = 10.73LEE487 pKa = 4.19QANVTKK493 pKa = 9.94RR494 pKa = 11.84TVGNIHH500 pKa = 6.59NGDD503 pKa = 3.51DD504 pKa = 3.49VYY506 pKa = 11.99ANIRR510 pKa = 11.84TIGDD514 pKa = 3.59ALDD517 pKa = 3.18IMRR520 pKa = 11.84DD521 pKa = 3.35AKK523 pKa = 11.03SIGVRR528 pKa = 11.84AQMTKK533 pKa = 10.09TNIGTIGEE541 pKa = 4.27FLRR544 pKa = 11.84IDD546 pKa = 3.84NKK548 pKa = 11.13AVDD551 pKa = 3.8ATGAQYY557 pKa = 9.18LTRR560 pKa = 11.84SCATAVHH567 pKa = 6.17GRR569 pKa = 11.84IEE571 pKa = 4.56SDD573 pKa = 3.46TPNSTRR579 pKa = 11.84SVVEE583 pKa = 3.57ANMIRR588 pKa = 11.84LEE590 pKa = 3.96ALKK593 pKa = 10.92ARR595 pKa = 11.84GGDD598 pKa = 3.56SNKK601 pKa = 10.16IKK603 pKa = 10.54VIKK606 pKa = 10.44DD607 pKa = 3.26NMISRR612 pKa = 11.84IARR615 pKa = 11.84KK616 pKa = 9.32HH617 pKa = 5.31DD618 pKa = 3.35TSEE621 pKa = 3.78EE622 pKa = 4.39TICDD626 pKa = 4.3LIEE629 pKa = 4.14TPKK632 pKa = 11.18ALGGLKK638 pKa = 10.1DD639 pKa = 4.23DD640 pKa = 4.78NKK642 pKa = 9.95PVPWRR647 pKa = 11.84LIIKK651 pKa = 10.5LEE653 pKa = 4.22GEE655 pKa = 4.51LDD657 pKa = 3.51SMAYY661 pKa = 9.87EE662 pKa = 4.93LANKK666 pKa = 7.91MKK668 pKa = 10.53KK669 pKa = 9.93AVNNYY674 pKa = 8.66HH675 pKa = 7.2KK676 pKa = 10.31AVCDD680 pKa = 3.59MLEE683 pKa = 4.02VEE685 pKa = 4.97MPTGYY690 pKa = 11.41DD691 pKa = 3.05MLNGINEE698 pKa = 4.19GMLQTKK704 pKa = 9.7KK705 pKa = 9.91MRR707 pKa = 11.84IEE709 pKa = 4.37KK710 pKa = 10.23IKK712 pKa = 10.52IDD714 pKa = 3.3IKK716 pKa = 10.64LAARR720 pKa = 11.84ICAMHH725 pKa = 6.6KK726 pKa = 10.19AWGGSGDD733 pKa = 3.65GGMAATMRR741 pKa = 11.84IFTDD745 pKa = 2.66IKK747 pKa = 8.12MHH749 pKa = 6.16RR750 pKa = 11.84KK751 pKa = 8.74HH752 pKa = 6.67RR753 pKa = 11.84RR754 pKa = 11.84LRR756 pKa = 11.84DD757 pKa = 3.26QVRR760 pKa = 11.84RR761 pKa = 11.84LVKK764 pKa = 10.76SGDD767 pKa = 3.2FNLWFNTVYY776 pKa = 11.07
MM1 pKa = 7.82LITVYY6 pKa = 10.77ILMGLRR12 pKa = 11.84TRR14 pKa = 11.84GAVDD18 pKa = 3.26EE19 pKa = 4.79ALLNVKK25 pKa = 9.57ALHH28 pKa = 6.45KK29 pKa = 10.8LHH31 pKa = 6.07GQYY34 pKa = 11.01VLAIPVLFGNVIAYY48 pKa = 9.61YY49 pKa = 10.23VDD51 pKa = 3.15KK52 pKa = 10.99HH53 pKa = 6.43DD54 pKa = 3.84VTMADD59 pKa = 3.36KK60 pKa = 10.74KK61 pKa = 9.99VAKK64 pKa = 9.89QAGLVIANNVFVDD77 pKa = 4.74FCPDD81 pKa = 2.84ARR83 pKa = 11.84VDD85 pKa = 3.84YY86 pKa = 10.22IAVMGNMAKK95 pKa = 10.36GLHH98 pKa = 6.04KK99 pKa = 9.66KK100 pKa = 7.74TRR102 pKa = 11.84INDD105 pKa = 3.53RR106 pKa = 11.84EE107 pKa = 4.37LIRR110 pKa = 11.84SRR112 pKa = 11.84ISQRR116 pKa = 11.84HH117 pKa = 4.8HH118 pKa = 4.37MHH120 pKa = 6.46IKK122 pKa = 10.42VGMLDD127 pKa = 3.57SEE129 pKa = 4.33GDD131 pKa = 3.59EE132 pKa = 4.96NEE134 pKa = 4.16IEE136 pKa = 4.01LDD138 pKa = 3.75GVKK141 pKa = 10.43GYY143 pKa = 10.65IMHH146 pKa = 7.71RR147 pKa = 11.84LMKK150 pKa = 8.33MTTVRR155 pKa = 11.84QTVTTFLLYY164 pKa = 10.77VEE166 pKa = 5.6LAPSWAADD174 pKa = 3.41VLAVIVSRR182 pKa = 11.84HH183 pKa = 5.1SEE185 pKa = 3.67WAQLMDD191 pKa = 4.26DD192 pKa = 4.49LKK194 pKa = 11.09LFSMYY199 pKa = 10.01VKK201 pKa = 10.35QYY203 pKa = 8.45QHH205 pKa = 6.85VIRR208 pKa = 11.84SDD210 pKa = 3.53LAVLFEE216 pKa = 5.2LNVLANRR223 pKa = 11.84SDD225 pKa = 4.94SEE227 pKa = 4.22MDD229 pKa = 2.96WAADD233 pKa = 3.0IAKK236 pKa = 8.28RR237 pKa = 11.84TKK239 pKa = 10.68SVVATPDD246 pKa = 3.07QEE248 pKa = 4.24RR249 pKa = 11.84LYY251 pKa = 10.8RR252 pKa = 11.84DD253 pKa = 3.66GVSMFTSARR262 pKa = 11.84AEE264 pKa = 4.22GKK266 pKa = 10.13QPVTMKK272 pKa = 9.93WEE274 pKa = 4.57EE275 pKa = 4.0YY276 pKa = 10.98RR277 pKa = 11.84NMTWGLTPPGAVHH290 pKa = 6.61SPEE293 pKa = 3.71GDD295 pKa = 3.14FRR297 pKa = 11.84RR298 pKa = 11.84NISTIPSKK306 pKa = 10.83LRR308 pKa = 11.84NKK310 pKa = 8.61KK311 pKa = 8.27TYY313 pKa = 10.79NSVFRR318 pKa = 11.84PKK320 pKa = 10.26EE321 pKa = 3.79LKK323 pKa = 10.2HH324 pKa = 7.45YY325 pKa = 7.33MAQRR329 pKa = 11.84PKK331 pKa = 10.3IIAKK335 pKa = 10.06PSVKK339 pKa = 10.66YY340 pKa = 9.97EE341 pKa = 3.7WGKK344 pKa = 9.93NRR346 pKa = 11.84ALYY349 pKa = 10.66GCDD352 pKa = 3.4LVSHH356 pKa = 6.54MMTDD360 pKa = 3.87FGMMQAEE367 pKa = 4.39STLPSWMPTGRR378 pKa = 11.84MADD381 pKa = 3.48EE382 pKa = 4.27NRR384 pKa = 11.84VKK386 pKa = 10.68RR387 pKa = 11.84QIATMGNGIPFCYY400 pKa = 10.51DD401 pKa = 2.8FDD403 pKa = 4.94DD404 pKa = 4.89FNSQHH409 pKa = 5.81SVKK412 pKa = 10.75SMQTVITAWYY422 pKa = 9.13SVYY425 pKa = 11.08GDD427 pKa = 5.26LISDD431 pKa = 3.84EE432 pKa = 4.44QKK434 pKa = 11.15LCVDD438 pKa = 4.04WVVQSLEE445 pKa = 3.92TQLIINSVGLGSSKK459 pKa = 10.72YY460 pKa = 9.92IEE462 pKa = 3.96VDD464 pKa = 3.04GTLFSGWRR472 pKa = 11.84LTSFMNTCLNYY483 pKa = 10.7LYY485 pKa = 10.73LEE487 pKa = 4.19QANVTKK493 pKa = 9.94RR494 pKa = 11.84TVGNIHH500 pKa = 6.59NGDD503 pKa = 3.51DD504 pKa = 3.49VYY506 pKa = 11.99ANIRR510 pKa = 11.84TIGDD514 pKa = 3.59ALDD517 pKa = 3.18IMRR520 pKa = 11.84DD521 pKa = 3.35AKK523 pKa = 11.03SIGVRR528 pKa = 11.84AQMTKK533 pKa = 10.09TNIGTIGEE541 pKa = 4.27FLRR544 pKa = 11.84IDD546 pKa = 3.84NKK548 pKa = 11.13AVDD551 pKa = 3.8ATGAQYY557 pKa = 9.18LTRR560 pKa = 11.84SCATAVHH567 pKa = 6.17GRR569 pKa = 11.84IEE571 pKa = 4.56SDD573 pKa = 3.46TPNSTRR579 pKa = 11.84SVVEE583 pKa = 3.57ANMIRR588 pKa = 11.84LEE590 pKa = 3.96ALKK593 pKa = 10.92ARR595 pKa = 11.84GGDD598 pKa = 3.56SNKK601 pKa = 10.16IKK603 pKa = 10.54VIKK606 pKa = 10.44DD607 pKa = 3.26NMISRR612 pKa = 11.84IARR615 pKa = 11.84KK616 pKa = 9.32HH617 pKa = 5.31DD618 pKa = 3.35TSEE621 pKa = 3.78EE622 pKa = 4.39TICDD626 pKa = 4.3LIEE629 pKa = 4.14TPKK632 pKa = 11.18ALGGLKK638 pKa = 10.1DD639 pKa = 4.23DD640 pKa = 4.78NKK642 pKa = 9.95PVPWRR647 pKa = 11.84LIIKK651 pKa = 10.5LEE653 pKa = 4.22GEE655 pKa = 4.51LDD657 pKa = 3.51SMAYY661 pKa = 9.87EE662 pKa = 4.93LANKK666 pKa = 7.91MKK668 pKa = 10.53KK669 pKa = 9.93AVNNYY674 pKa = 8.66HH675 pKa = 7.2KK676 pKa = 10.31AVCDD680 pKa = 3.59MLEE683 pKa = 4.02VEE685 pKa = 4.97MPTGYY690 pKa = 11.41DD691 pKa = 3.05MLNGINEE698 pKa = 4.19GMLQTKK704 pKa = 9.7KK705 pKa = 9.91MRR707 pKa = 11.84IEE709 pKa = 4.37KK710 pKa = 10.23IKK712 pKa = 10.52IDD714 pKa = 3.3IKK716 pKa = 10.64LAARR720 pKa = 11.84ICAMHH725 pKa = 6.6KK726 pKa = 10.19AWGGSGDD733 pKa = 3.65GGMAATMRR741 pKa = 11.84IFTDD745 pKa = 2.66IKK747 pKa = 8.12MHH749 pKa = 6.16RR750 pKa = 11.84KK751 pKa = 8.74HH752 pKa = 6.67RR753 pKa = 11.84RR754 pKa = 11.84LRR756 pKa = 11.84DD757 pKa = 3.26QVRR760 pKa = 11.84RR761 pKa = 11.84LVKK764 pKa = 10.76SGDD767 pKa = 3.2FNLWFNTVYY776 pKa = 11.07
Molecular weight: 87.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1356 |
580 |
776 |
678.0 |
77.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.932 ± 0.477 | 1.18 ± 0.018 |
7.153 ± 0.285 | 5.383 ± 0.542 |
3.171 ± 0.636 | 6.047 ± 0.235 |
2.581 ± 0.11 | 6.121 ± 0.511 |
6.268 ± 0.948 | 8.776 ± 0.692 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.499 ± 0.691 | 4.794 ± 0.091 |
3.171 ± 0.523 | 2.876 ± 0.036 |
5.9 ± 0.479 | 5.973 ± 0.381 |
6.195 ± 0.235 | 7.227 ± 0.331 |
1.549 ± 0.111 | 4.204 ± 0.638 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
