
Torilis crimson leaf virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins
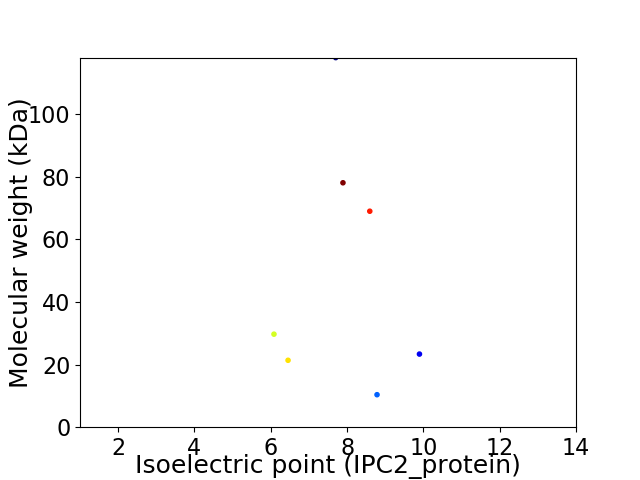
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
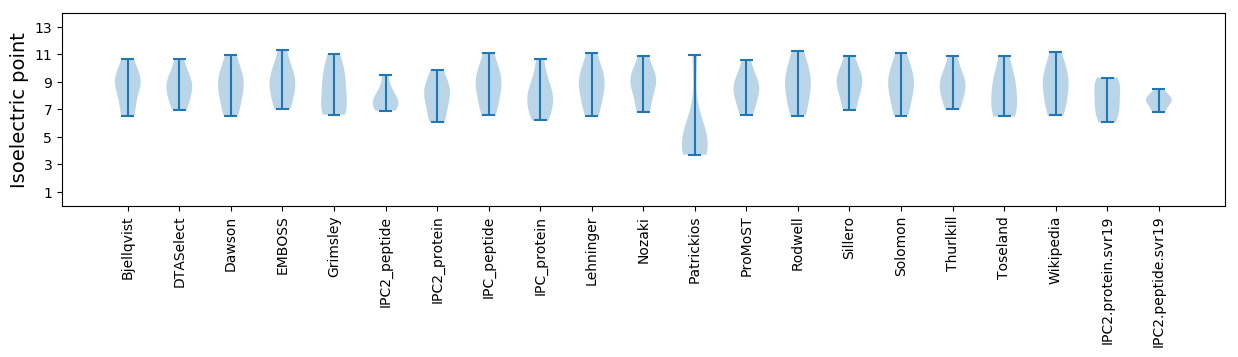
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6H2MW64|A0A6H2MW64_9LUTE Readthrough protein OS=Torilis crimson leaf virus OX=1862685 PE=3 SV=1
MM1 pKa = 7.21QLEE4 pKa = 4.29IQRR7 pKa = 11.84SGIVTFTARR16 pKa = 11.84SFFSRR21 pKa = 11.84LFAYY25 pKa = 9.91RR26 pKa = 11.84YY27 pKa = 9.61YY28 pKa = 9.28ITHH31 pKa = 7.61ALTLSRR37 pKa = 11.84QLNNVEE43 pKa = 3.95PSFPEE48 pKa = 3.74TLPRR52 pKa = 11.84SLLFSLPFIINNNYY66 pKa = 10.85DD67 pKa = 3.14MGDD70 pKa = 3.35GYY72 pKa = 10.77IRR74 pKa = 11.84FQPDD78 pKa = 2.88HH79 pKa = 6.78LRR81 pKa = 11.84EE82 pKa = 3.88FVLWGLICGYY92 pKa = 9.07YY93 pKa = 9.76PRR95 pKa = 11.84LNKK98 pKa = 9.93RR99 pKa = 11.84GRR101 pKa = 11.84RR102 pKa = 11.84GYY104 pKa = 10.34RR105 pKa = 11.84ATLAARR111 pKa = 11.84VEE113 pKa = 4.0RR114 pKa = 11.84RR115 pKa = 11.84AYY117 pKa = 10.86LSTLHH122 pKa = 6.49NLDD125 pKa = 2.99WRR127 pKa = 11.84NLEE130 pKa = 3.86EE131 pKa = 4.39NMFRR135 pKa = 11.84RR136 pKa = 11.84QEE138 pKa = 4.15CFFQSAEE145 pKa = 4.04GFSRR149 pKa = 11.84MLGGLFSIWNRR160 pKa = 11.84DD161 pKa = 2.85AANRR165 pKa = 11.84YY166 pKa = 9.71GEE168 pKa = 4.69LPLDD172 pKa = 3.7ACRR175 pKa = 11.84NLVDD179 pKa = 4.79SIFVGSKK186 pKa = 10.37VYY188 pKa = 10.12MDD190 pKa = 5.07LSDD193 pKa = 5.22LFHH196 pKa = 7.11DD197 pKa = 4.21TCPCIGYY204 pKa = 8.92SHH206 pKa = 7.3LNNNGDD212 pKa = 4.07LPCGQMDD219 pKa = 5.78LFTAPYY225 pKa = 10.91ALYY228 pKa = 9.6TLSADD233 pKa = 3.22HH234 pKa = 6.82HH235 pKa = 6.85KK236 pKa = 9.95IHH238 pKa = 6.64HH239 pKa = 6.17WNAGTSEE246 pKa = 4.27EE247 pKa = 4.25NSDD250 pKa = 4.59SEE252 pKa = 5.36GEE254 pKa = 4.29GLL256 pKa = 4.7
MM1 pKa = 7.21QLEE4 pKa = 4.29IQRR7 pKa = 11.84SGIVTFTARR16 pKa = 11.84SFFSRR21 pKa = 11.84LFAYY25 pKa = 9.91RR26 pKa = 11.84YY27 pKa = 9.61YY28 pKa = 9.28ITHH31 pKa = 7.61ALTLSRR37 pKa = 11.84QLNNVEE43 pKa = 3.95PSFPEE48 pKa = 3.74TLPRR52 pKa = 11.84SLLFSLPFIINNNYY66 pKa = 10.85DD67 pKa = 3.14MGDD70 pKa = 3.35GYY72 pKa = 10.77IRR74 pKa = 11.84FQPDD78 pKa = 2.88HH79 pKa = 6.78LRR81 pKa = 11.84EE82 pKa = 3.88FVLWGLICGYY92 pKa = 9.07YY93 pKa = 9.76PRR95 pKa = 11.84LNKK98 pKa = 9.93RR99 pKa = 11.84GRR101 pKa = 11.84RR102 pKa = 11.84GYY104 pKa = 10.34RR105 pKa = 11.84ATLAARR111 pKa = 11.84VEE113 pKa = 4.0RR114 pKa = 11.84RR115 pKa = 11.84AYY117 pKa = 10.86LSTLHH122 pKa = 6.49NLDD125 pKa = 2.99WRR127 pKa = 11.84NLEE130 pKa = 3.86EE131 pKa = 4.39NMFRR135 pKa = 11.84RR136 pKa = 11.84QEE138 pKa = 4.15CFFQSAEE145 pKa = 4.04GFSRR149 pKa = 11.84MLGGLFSIWNRR160 pKa = 11.84DD161 pKa = 2.85AANRR165 pKa = 11.84YY166 pKa = 9.71GEE168 pKa = 4.69LPLDD172 pKa = 3.7ACRR175 pKa = 11.84NLVDD179 pKa = 4.79SIFVGSKK186 pKa = 10.37VYY188 pKa = 10.12MDD190 pKa = 5.07LSDD193 pKa = 5.22LFHH196 pKa = 7.11DD197 pKa = 4.21TCPCIGYY204 pKa = 8.92SHH206 pKa = 7.3LNNNGDD212 pKa = 4.07LPCGQMDD219 pKa = 5.78LFTAPYY225 pKa = 10.91ALYY228 pKa = 9.6TLSADD233 pKa = 3.22HH234 pKa = 6.82HH235 pKa = 6.85KK236 pKa = 9.95IHH238 pKa = 6.64HH239 pKa = 6.17WNAGTSEE246 pKa = 4.27EE247 pKa = 4.25NSDD250 pKa = 4.59SEE252 pKa = 5.36GEE254 pKa = 4.29GLL256 pKa = 4.7
Molecular weight: 29.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6H2MW59|A0A6H2MW59_9LUTE P0 OS=Torilis crimson leaf virus OX=1862685 PE=4 SV=1
MM1 pKa = 7.98PIYY4 pKa = 10.82GLTKK8 pKa = 10.05RR9 pKa = 11.84STGNKK14 pKa = 8.7PMVVVNSPPRR24 pKa = 11.84RR25 pKa = 11.84PRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84VRR31 pKa = 11.84TKK33 pKa = 10.3PIVVVQTTQPGRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84GRR53 pKa = 11.84NGRR56 pKa = 11.84GPRR59 pKa = 11.84TMGQSRR65 pKa = 11.84GGANRR70 pKa = 11.84EE71 pKa = 3.63QLVFSKK77 pKa = 11.03DD78 pKa = 3.69DD79 pKa = 3.42IKK81 pKa = 11.46GNSYY85 pKa = 10.83GAITFGKK92 pKa = 10.08DD93 pKa = 3.64LSDD96 pKa = 3.86HH97 pKa = 6.77PAFSSGILKK106 pKa = 10.4AFHH109 pKa = 6.93EE110 pKa = 4.46YY111 pKa = 10.6KK112 pKa = 9.35ITNLRR117 pKa = 11.84VIFKK121 pKa = 10.49SEE123 pKa = 3.96APSTTGGSLAYY134 pKa = 9.74EE135 pKa = 4.61LDD137 pKa = 3.63PHH139 pKa = 7.38CEE141 pKa = 4.02LTSLSSKK148 pKa = 9.75IYY150 pKa = 10.65KK151 pKa = 10.54FGATKK156 pKa = 10.42GGQKK160 pKa = 8.26TWTAKK165 pKa = 10.18EE166 pKa = 3.82INGTEE171 pKa = 3.79WHH173 pKa = 6.71SSSEE177 pKa = 3.96NQFRR181 pKa = 11.84LLYY184 pKa = 10.08KK185 pKa = 10.91GNGDD189 pKa = 3.48STQIGSFEE197 pKa = 3.51IHH199 pKa = 5.26YY200 pKa = 8.84TVLFQNPKK208 pKa = 9.88
MM1 pKa = 7.98PIYY4 pKa = 10.82GLTKK8 pKa = 10.05RR9 pKa = 11.84STGNKK14 pKa = 8.7PMVVVNSPPRR24 pKa = 11.84RR25 pKa = 11.84PRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84VRR31 pKa = 11.84TKK33 pKa = 10.3PIVVVQTTQPGRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84GRR53 pKa = 11.84NGRR56 pKa = 11.84GPRR59 pKa = 11.84TMGQSRR65 pKa = 11.84GGANRR70 pKa = 11.84EE71 pKa = 3.63QLVFSKK77 pKa = 11.03DD78 pKa = 3.69DD79 pKa = 3.42IKK81 pKa = 11.46GNSYY85 pKa = 10.83GAITFGKK92 pKa = 10.08DD93 pKa = 3.64LSDD96 pKa = 3.86HH97 pKa = 6.77PAFSSGILKK106 pKa = 10.4AFHH109 pKa = 6.93EE110 pKa = 4.46YY111 pKa = 10.6KK112 pKa = 9.35ITNLRR117 pKa = 11.84VIFKK121 pKa = 10.49SEE123 pKa = 3.96APSTTGGSLAYY134 pKa = 9.74EE135 pKa = 4.61LDD137 pKa = 3.63PHH139 pKa = 7.38CEE141 pKa = 4.02LTSLSSKK148 pKa = 9.75IYY150 pKa = 10.65KK151 pKa = 10.54FGATKK156 pKa = 10.42GGQKK160 pKa = 8.26TWTAKK165 pKa = 10.18EE166 pKa = 3.82INGTEE171 pKa = 3.79WHH173 pKa = 6.71SSSEE177 pKa = 3.96NQFRR181 pKa = 11.84LLYY184 pKa = 10.08KK185 pKa = 10.91GNGDD189 pKa = 3.48STQIGSFEE197 pKa = 3.51IHH199 pKa = 5.26YY200 pKa = 8.84TVLFQNPKK208 pKa = 9.88
Molecular weight: 23.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3123 |
92 |
1051 |
446.1 |
49.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.116 ± 0.47 | 1.569 ± 0.254 |
4.131 ± 0.529 | 6.084 ± 0.356 |
4.387 ± 0.365 | 6.948 ± 0.556 |
1.921 ± 0.209 | 5.027 ± 0.391 |
5.828 ± 0.694 | 9.03 ± 0.541 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.985 ± 0.161 | 5.379 ± 0.28 |
5.476 ± 0.343 | 3.746 ± 0.424 |
6.308 ± 0.798 | 8.774 ± 0.459 |
7.077 ± 0.527 | 5.572 ± 0.499 |
1.697 ± 0.179 | 2.914 ± 0.494 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
