
Auraticoccus cholistanensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Auraticoccus
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
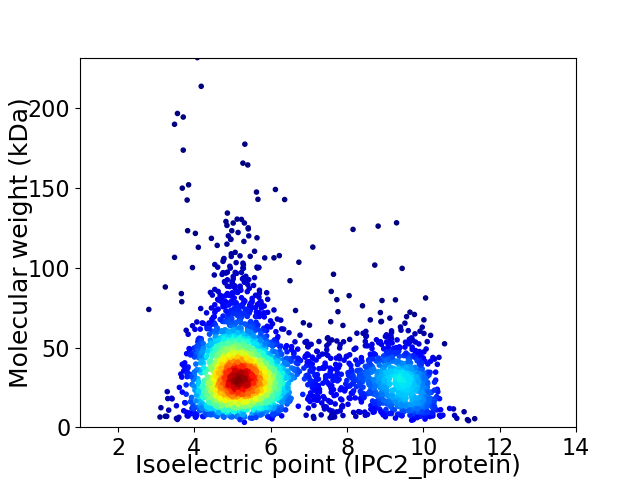
Virtual 2D-PAGE plot for 3358 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
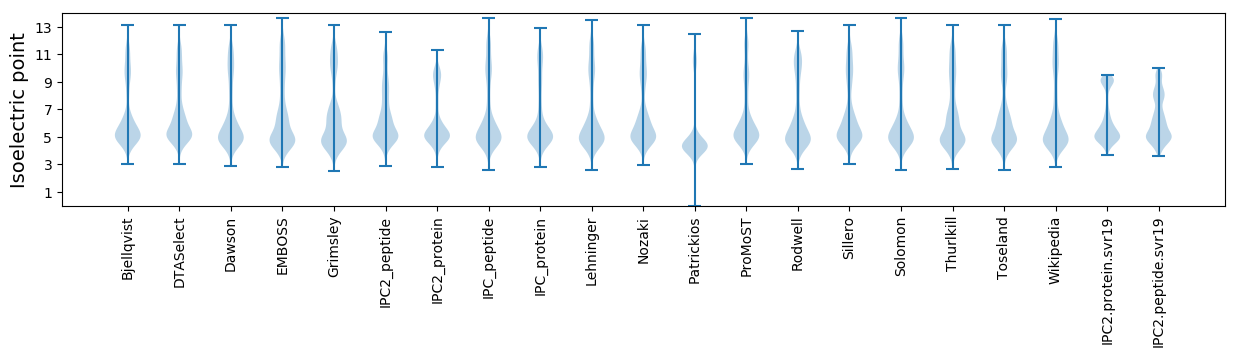
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6A9UWB0|A0A6A9UWB0_9ACTN Fe-S cluster assembly ATPase SufC OS=Auraticoccus cholistanensis OX=2656650 GN=sufC PE=3 SV=1
MM1 pKa = 7.18TALFMSMFSAGVAHH15 pKa = 7.47AATVYY20 pKa = 10.13EE21 pKa = 5.62IEE23 pKa = 4.67GEE25 pKa = 4.26WEE27 pKa = 4.02AGTPTTVGRR36 pKa = 11.84GDD38 pKa = 3.64VVTGIWRR45 pKa = 11.84VNVNDD50 pKa = 4.83DD51 pKa = 4.1AEE53 pKa = 5.16APANDD58 pKa = 3.62PVEE61 pKa = 4.26NVLFSVTLDD70 pKa = 3.01NGVFRR75 pKa = 11.84EE76 pKa = 4.25LPDD79 pKa = 3.3VCVTGDD85 pKa = 3.51EE86 pKa = 4.4VDD88 pKa = 3.82PQSTLSEE95 pKa = 4.47DD96 pKa = 4.02NRR98 pKa = 11.84TLTCNLGTVEE108 pKa = 4.04QGSAVVIQTPVVVDD122 pKa = 3.96GLTGEE127 pKa = 4.09QLTAVGEE134 pKa = 4.46INGQEE139 pKa = 4.19QPLPPLNIVNEE150 pKa = 4.33FGMDD154 pKa = 3.07IQFGQNTGSYY164 pKa = 8.69YY165 pKa = 10.28WNDD168 pKa = 4.25DD169 pKa = 3.51YY170 pKa = 11.81TQLDD174 pKa = 3.28VDD176 pKa = 5.12LNWSLRR182 pKa = 11.84LAAGSDD188 pKa = 3.69PGPDD192 pKa = 2.87SVTYY196 pKa = 10.53RR197 pKa = 11.84LQLTEE202 pKa = 4.32TNNGTIEE209 pKa = 4.78PGTGLYY215 pKa = 10.74GDD217 pKa = 4.42VGCGPFNAYY226 pKa = 8.78IANGHH231 pKa = 6.32PWSEE235 pKa = 4.11LPNYY239 pKa = 9.46PADD242 pKa = 3.72QQTSFVDD249 pKa = 3.52SCTLTPVAGQPGLFDD264 pKa = 3.79LTLDD268 pKa = 4.71GINYY272 pKa = 10.06DD273 pKa = 3.87LATVPTKK280 pKa = 10.95DD281 pKa = 3.21SAGNPLPTDD290 pKa = 3.45WSYY293 pKa = 11.47IASGSLWFSITTNRR307 pKa = 11.84AGSIALQTNAPTYY320 pKa = 10.51EE321 pKa = 4.12STTGLFSTDD330 pKa = 2.81LTEE333 pKa = 5.32NNTTTKK339 pKa = 10.62SYY341 pKa = 9.11TLPGRR346 pKa = 11.84WSAPWYY352 pKa = 9.7RR353 pKa = 11.84GHH355 pKa = 6.77TGAGGTNWDD364 pKa = 3.11DD365 pKa = 3.79TYY367 pKa = 11.22RR368 pKa = 11.84VSAGQTVAQFVNNTASRR385 pKa = 11.84LGDD388 pKa = 3.46VPDD391 pKa = 3.87EE392 pKa = 4.38QIHH395 pKa = 5.1GQCLVFDD402 pKa = 4.47TAYY405 pKa = 10.76SSYY408 pKa = 11.1ASPEE412 pKa = 4.13GEE414 pKa = 4.08RR415 pKa = 11.84VPMIRR420 pKa = 11.84GYY422 pKa = 9.96DD423 pKa = 3.36QEE425 pKa = 4.75GAGGAAEE432 pKa = 4.74LEE434 pKa = 4.19NPPTLEE440 pKa = 4.23YY441 pKa = 11.1YY442 pKa = 10.69VGTVTDD448 pKa = 4.67PNAFVCGNDD457 pKa = 3.21PGNWVDD463 pKa = 5.22EE464 pKa = 4.6EE465 pKa = 4.19PADD468 pKa = 4.19LEE470 pKa = 4.37TVQAVRR476 pKa = 11.84ILYY479 pKa = 7.83PHH481 pKa = 6.72SLLAEE486 pKa = 4.43EE487 pKa = 5.52GWQGIQLLAYY497 pKa = 9.3TEE499 pKa = 4.89LDD501 pKa = 3.52DD502 pKa = 6.08DD503 pKa = 5.19LEE505 pKa = 5.74IGQDD509 pKa = 2.68VWMFGSVFANGVWEE523 pKa = 4.89GPGEE527 pKa = 4.01PWDD530 pKa = 4.18SNVITPTPGTRR541 pKa = 11.84YY542 pKa = 8.49PATNGRR548 pKa = 11.84RR549 pKa = 11.84DD550 pKa = 3.43ILRR553 pKa = 11.84IIYY556 pKa = 7.52ATPAISKK563 pKa = 10.55AGDD566 pKa = 3.49RR567 pKa = 11.84QVVRR571 pKa = 11.84PGEE574 pKa = 4.09PATFTLTYY582 pKa = 9.51SANGAGAIPDD592 pKa = 3.91TVDD595 pKa = 2.97GYY597 pKa = 11.17EE598 pKa = 4.28IVDD601 pKa = 3.78TLPVGMTYY609 pKa = 10.33VPEE612 pKa = 4.24STEE615 pKa = 3.95PEE617 pKa = 4.22PEE619 pKa = 3.84VTTNGEE625 pKa = 4.13GQQVLTWTLNEE636 pKa = 4.37VPTNTPNTLTYY647 pKa = 10.28QAVASDD653 pKa = 4.1SVTPGQTLTNTATSSLAGQSSRR675 pKa = 11.84PASAQVTVASSGYY688 pKa = 7.86TLLGKK693 pKa = 8.54STAQWFMNNPDD704 pKa = 3.67GEE706 pKa = 4.69GNSTGSWTVNLRR718 pKa = 11.84SSDD721 pKa = 4.35PLPQAYY727 pKa = 8.55TDD729 pKa = 4.17TIDD732 pKa = 3.21ILPYY736 pKa = 10.53NGDD739 pKa = 3.05GRR741 pKa = 11.84GTDD744 pKa = 3.73FQGTYY749 pKa = 10.58SVTSVDD755 pKa = 3.75APAGATVYY763 pKa = 8.88YY764 pKa = 6.93TTADD768 pKa = 4.0RR769 pKa = 11.84EE770 pKa = 4.42TLSDD774 pKa = 4.53DD775 pKa = 3.94PADD778 pKa = 4.9EE779 pKa = 4.94INGAPNAPSDD789 pKa = 3.32IWSTTPVANPTAIRR803 pKa = 11.84VIGGQLNPGEE813 pKa = 4.6AFAFDD818 pKa = 3.79VNIATEE824 pKa = 4.59GAAPGDD830 pKa = 3.49VWVNRR835 pKa = 11.84AQSRR839 pKa = 11.84AEE841 pKa = 3.74HH842 pKa = 5.91TRR844 pKa = 11.84LVMRR848 pKa = 11.84TSEE851 pKa = 4.44PLTMGTMYY859 pKa = 10.69SASLKK864 pKa = 10.62KK865 pKa = 10.48YY866 pKa = 8.48VQDD869 pKa = 5.7RR870 pKa = 11.84DD871 pKa = 4.47GEE873 pKa = 4.18WHH875 pKa = 7.18DD876 pKa = 4.27AQDD879 pKa = 3.57ASDD882 pKa = 4.14YY883 pKa = 10.51PVFRR887 pKa = 11.84IGDD890 pKa = 3.73EE891 pKa = 3.77VTYY894 pKa = 10.6RR895 pKa = 11.84IGVTNTGQGTLTNVQVTDD913 pKa = 3.83DD914 pKa = 4.85KK915 pKa = 11.35FDD917 pKa = 3.27QGAFLIEE924 pKa = 4.04EE925 pKa = 4.59LAPGDD930 pKa = 3.98VEE932 pKa = 4.16YY933 pKa = 11.06HH934 pKa = 6.77EE935 pKa = 4.35YY936 pKa = 10.06TVTIEE941 pKa = 4.5DD942 pKa = 3.54SFGTSFVNTASATADD957 pKa = 3.43QPEE960 pKa = 4.38DD961 pKa = 3.88AEE963 pKa = 5.5DD964 pKa = 3.7PVQINSDD971 pKa = 3.67PAGVEE976 pKa = 3.86VANYY980 pKa = 10.46SVVKK984 pKa = 10.18TSDD987 pKa = 3.22PSGAAVNPGDD997 pKa = 3.63VVTYY1001 pKa = 8.55TVEE1004 pKa = 4.13VTQEE1008 pKa = 3.73GSVAAEE1014 pKa = 3.92ATFSDD1019 pKa = 4.56DD1020 pKa = 5.16LSDD1023 pKa = 3.94VLDD1026 pKa = 4.81DD1027 pKa = 4.0ATVDD1031 pKa = 3.33ADD1033 pKa = 4.58SIVASTGTAEE1043 pKa = 4.59LDD1045 pKa = 3.53GEE1047 pKa = 4.34ALVWTGTVPVGEE1059 pKa = 4.54VATVTYY1065 pKa = 9.59EE1066 pKa = 4.16AVVKK1070 pKa = 10.67SYY1072 pKa = 11.62DD1073 pKa = 3.72EE1074 pKa = 4.5IEE1076 pKa = 4.91ADD1078 pKa = 4.64GNWILDD1084 pKa = 3.14NVVFSEE1090 pKa = 4.77GCAEE1094 pKa = 4.59EE1095 pKa = 5.58GDD1097 pKa = 4.47CNPPEE1102 pKa = 4.04NPIGKK1107 pKa = 8.33FTYY1110 pKa = 10.45SKK1112 pKa = 11.1VSDD1115 pKa = 4.6PEE1117 pKa = 4.61PGTAVAPGQEE1127 pKa = 3.75IDD1129 pKa = 3.52YY1130 pKa = 8.81TITVSQQGEE1139 pKa = 3.97AAIPGASVVDD1149 pKa = 4.3DD1150 pKa = 4.73LSDD1153 pKa = 3.72VLDD1156 pKa = 5.02DD1157 pKa = 4.13ATLDD1161 pKa = 3.75PATISASAGEE1171 pKa = 4.08AVLDD1175 pKa = 4.31GSTLSWTGDD1184 pKa = 3.68LAVDD1188 pKa = 3.75DD1189 pKa = 4.91VVTITYY1195 pKa = 8.13TVVVVEE1201 pKa = 4.96DD1202 pKa = 4.45GSDD1205 pKa = 3.39EE1206 pKa = 4.16LVNPVTSEE1214 pKa = 4.12DD1215 pKa = 3.6PRR1217 pKa = 11.84GTCDD1221 pKa = 3.6EE1222 pKa = 4.28EE1223 pKa = 4.78VGCEE1227 pKa = 3.88TTHH1230 pKa = 6.91LKK1232 pKa = 10.71GHH1234 pKa = 5.74FTYY1237 pKa = 10.91SKK1239 pKa = 9.45TADD1242 pKa = 3.87PGSGATVAEE1251 pKa = 4.44GDD1253 pKa = 3.96VITYY1257 pKa = 7.47TVTVSHH1263 pKa = 6.27QGKK1266 pKa = 10.09APLEE1270 pKa = 4.48DD1271 pKa = 3.25ATVVDD1276 pKa = 5.22DD1277 pKa = 4.3LTDD1280 pKa = 4.07VIDD1283 pKa = 4.01DD1284 pKa = 5.24AIYY1287 pKa = 11.0NGDD1290 pKa = 3.52VEE1292 pKa = 4.47ASSGAVRR1299 pKa = 11.84YY1300 pKa = 9.76ADD1302 pKa = 5.38SKK1304 pKa = 10.16LTWTGDD1310 pKa = 3.61LEE1312 pKa = 4.52VGQVVTFTYY1321 pKa = 9.55TVEE1324 pKa = 4.17VTGDD1328 pKa = 3.33GSDD1331 pKa = 3.42NLVNPVTTDD1340 pKa = 3.57DD1341 pKa = 4.03PRR1343 pKa = 11.84GTCDD1347 pKa = 3.61EE1348 pKa = 4.38AVGCQTEE1355 pKa = 4.72HH1356 pKa = 7.05PLEE1359 pKa = 4.68DD1360 pKa = 4.02APVPAPGLGGGGTGVGGNLPNTGAPAVLGTLLAGLLMVGAGLTVVARR1407 pKa = 11.84RR1408 pKa = 11.84RR1409 pKa = 11.84RR1410 pKa = 11.84ISLGKK1415 pKa = 10.25GQTSSLL1421 pKa = 3.73
MM1 pKa = 7.18TALFMSMFSAGVAHH15 pKa = 7.47AATVYY20 pKa = 10.13EE21 pKa = 5.62IEE23 pKa = 4.67GEE25 pKa = 4.26WEE27 pKa = 4.02AGTPTTVGRR36 pKa = 11.84GDD38 pKa = 3.64VVTGIWRR45 pKa = 11.84VNVNDD50 pKa = 4.83DD51 pKa = 4.1AEE53 pKa = 5.16APANDD58 pKa = 3.62PVEE61 pKa = 4.26NVLFSVTLDD70 pKa = 3.01NGVFRR75 pKa = 11.84EE76 pKa = 4.25LPDD79 pKa = 3.3VCVTGDD85 pKa = 3.51EE86 pKa = 4.4VDD88 pKa = 3.82PQSTLSEE95 pKa = 4.47DD96 pKa = 4.02NRR98 pKa = 11.84TLTCNLGTVEE108 pKa = 4.04QGSAVVIQTPVVVDD122 pKa = 3.96GLTGEE127 pKa = 4.09QLTAVGEE134 pKa = 4.46INGQEE139 pKa = 4.19QPLPPLNIVNEE150 pKa = 4.33FGMDD154 pKa = 3.07IQFGQNTGSYY164 pKa = 8.69YY165 pKa = 10.28WNDD168 pKa = 4.25DD169 pKa = 3.51YY170 pKa = 11.81TQLDD174 pKa = 3.28VDD176 pKa = 5.12LNWSLRR182 pKa = 11.84LAAGSDD188 pKa = 3.69PGPDD192 pKa = 2.87SVTYY196 pKa = 10.53RR197 pKa = 11.84LQLTEE202 pKa = 4.32TNNGTIEE209 pKa = 4.78PGTGLYY215 pKa = 10.74GDD217 pKa = 4.42VGCGPFNAYY226 pKa = 8.78IANGHH231 pKa = 6.32PWSEE235 pKa = 4.11LPNYY239 pKa = 9.46PADD242 pKa = 3.72QQTSFVDD249 pKa = 3.52SCTLTPVAGQPGLFDD264 pKa = 3.79LTLDD268 pKa = 4.71GINYY272 pKa = 10.06DD273 pKa = 3.87LATVPTKK280 pKa = 10.95DD281 pKa = 3.21SAGNPLPTDD290 pKa = 3.45WSYY293 pKa = 11.47IASGSLWFSITTNRR307 pKa = 11.84AGSIALQTNAPTYY320 pKa = 10.51EE321 pKa = 4.12STTGLFSTDD330 pKa = 2.81LTEE333 pKa = 5.32NNTTTKK339 pKa = 10.62SYY341 pKa = 9.11TLPGRR346 pKa = 11.84WSAPWYY352 pKa = 9.7RR353 pKa = 11.84GHH355 pKa = 6.77TGAGGTNWDD364 pKa = 3.11DD365 pKa = 3.79TYY367 pKa = 11.22RR368 pKa = 11.84VSAGQTVAQFVNNTASRR385 pKa = 11.84LGDD388 pKa = 3.46VPDD391 pKa = 3.87EE392 pKa = 4.38QIHH395 pKa = 5.1GQCLVFDD402 pKa = 4.47TAYY405 pKa = 10.76SSYY408 pKa = 11.1ASPEE412 pKa = 4.13GEE414 pKa = 4.08RR415 pKa = 11.84VPMIRR420 pKa = 11.84GYY422 pKa = 9.96DD423 pKa = 3.36QEE425 pKa = 4.75GAGGAAEE432 pKa = 4.74LEE434 pKa = 4.19NPPTLEE440 pKa = 4.23YY441 pKa = 11.1YY442 pKa = 10.69VGTVTDD448 pKa = 4.67PNAFVCGNDD457 pKa = 3.21PGNWVDD463 pKa = 5.22EE464 pKa = 4.6EE465 pKa = 4.19PADD468 pKa = 4.19LEE470 pKa = 4.37TVQAVRR476 pKa = 11.84ILYY479 pKa = 7.83PHH481 pKa = 6.72SLLAEE486 pKa = 4.43EE487 pKa = 5.52GWQGIQLLAYY497 pKa = 9.3TEE499 pKa = 4.89LDD501 pKa = 3.52DD502 pKa = 6.08DD503 pKa = 5.19LEE505 pKa = 5.74IGQDD509 pKa = 2.68VWMFGSVFANGVWEE523 pKa = 4.89GPGEE527 pKa = 4.01PWDD530 pKa = 4.18SNVITPTPGTRR541 pKa = 11.84YY542 pKa = 8.49PATNGRR548 pKa = 11.84RR549 pKa = 11.84DD550 pKa = 3.43ILRR553 pKa = 11.84IIYY556 pKa = 7.52ATPAISKK563 pKa = 10.55AGDD566 pKa = 3.49RR567 pKa = 11.84QVVRR571 pKa = 11.84PGEE574 pKa = 4.09PATFTLTYY582 pKa = 9.51SANGAGAIPDD592 pKa = 3.91TVDD595 pKa = 2.97GYY597 pKa = 11.17EE598 pKa = 4.28IVDD601 pKa = 3.78TLPVGMTYY609 pKa = 10.33VPEE612 pKa = 4.24STEE615 pKa = 3.95PEE617 pKa = 4.22PEE619 pKa = 3.84VTTNGEE625 pKa = 4.13GQQVLTWTLNEE636 pKa = 4.37VPTNTPNTLTYY647 pKa = 10.28QAVASDD653 pKa = 4.1SVTPGQTLTNTATSSLAGQSSRR675 pKa = 11.84PASAQVTVASSGYY688 pKa = 7.86TLLGKK693 pKa = 8.54STAQWFMNNPDD704 pKa = 3.67GEE706 pKa = 4.69GNSTGSWTVNLRR718 pKa = 11.84SSDD721 pKa = 4.35PLPQAYY727 pKa = 8.55TDD729 pKa = 4.17TIDD732 pKa = 3.21ILPYY736 pKa = 10.53NGDD739 pKa = 3.05GRR741 pKa = 11.84GTDD744 pKa = 3.73FQGTYY749 pKa = 10.58SVTSVDD755 pKa = 3.75APAGATVYY763 pKa = 8.88YY764 pKa = 6.93TTADD768 pKa = 4.0RR769 pKa = 11.84EE770 pKa = 4.42TLSDD774 pKa = 4.53DD775 pKa = 3.94PADD778 pKa = 4.9EE779 pKa = 4.94INGAPNAPSDD789 pKa = 3.32IWSTTPVANPTAIRR803 pKa = 11.84VIGGQLNPGEE813 pKa = 4.6AFAFDD818 pKa = 3.79VNIATEE824 pKa = 4.59GAAPGDD830 pKa = 3.49VWVNRR835 pKa = 11.84AQSRR839 pKa = 11.84AEE841 pKa = 3.74HH842 pKa = 5.91TRR844 pKa = 11.84LVMRR848 pKa = 11.84TSEE851 pKa = 4.44PLTMGTMYY859 pKa = 10.69SASLKK864 pKa = 10.62KK865 pKa = 10.48YY866 pKa = 8.48VQDD869 pKa = 5.7RR870 pKa = 11.84DD871 pKa = 4.47GEE873 pKa = 4.18WHH875 pKa = 7.18DD876 pKa = 4.27AQDD879 pKa = 3.57ASDD882 pKa = 4.14YY883 pKa = 10.51PVFRR887 pKa = 11.84IGDD890 pKa = 3.73EE891 pKa = 3.77VTYY894 pKa = 10.6RR895 pKa = 11.84IGVTNTGQGTLTNVQVTDD913 pKa = 3.83DD914 pKa = 4.85KK915 pKa = 11.35FDD917 pKa = 3.27QGAFLIEE924 pKa = 4.04EE925 pKa = 4.59LAPGDD930 pKa = 3.98VEE932 pKa = 4.16YY933 pKa = 11.06HH934 pKa = 6.77EE935 pKa = 4.35YY936 pKa = 10.06TVTIEE941 pKa = 4.5DD942 pKa = 3.54SFGTSFVNTASATADD957 pKa = 3.43QPEE960 pKa = 4.38DD961 pKa = 3.88AEE963 pKa = 5.5DD964 pKa = 3.7PVQINSDD971 pKa = 3.67PAGVEE976 pKa = 3.86VANYY980 pKa = 10.46SVVKK984 pKa = 10.18TSDD987 pKa = 3.22PSGAAVNPGDD997 pKa = 3.63VVTYY1001 pKa = 8.55TVEE1004 pKa = 4.13VTQEE1008 pKa = 3.73GSVAAEE1014 pKa = 3.92ATFSDD1019 pKa = 4.56DD1020 pKa = 5.16LSDD1023 pKa = 3.94VLDD1026 pKa = 4.81DD1027 pKa = 4.0ATVDD1031 pKa = 3.33ADD1033 pKa = 4.58SIVASTGTAEE1043 pKa = 4.59LDD1045 pKa = 3.53GEE1047 pKa = 4.34ALVWTGTVPVGEE1059 pKa = 4.54VATVTYY1065 pKa = 9.59EE1066 pKa = 4.16AVVKK1070 pKa = 10.67SYY1072 pKa = 11.62DD1073 pKa = 3.72EE1074 pKa = 4.5IEE1076 pKa = 4.91ADD1078 pKa = 4.64GNWILDD1084 pKa = 3.14NVVFSEE1090 pKa = 4.77GCAEE1094 pKa = 4.59EE1095 pKa = 5.58GDD1097 pKa = 4.47CNPPEE1102 pKa = 4.04NPIGKK1107 pKa = 8.33FTYY1110 pKa = 10.45SKK1112 pKa = 11.1VSDD1115 pKa = 4.6PEE1117 pKa = 4.61PGTAVAPGQEE1127 pKa = 3.75IDD1129 pKa = 3.52YY1130 pKa = 8.81TITVSQQGEE1139 pKa = 3.97AAIPGASVVDD1149 pKa = 4.3DD1150 pKa = 4.73LSDD1153 pKa = 3.72VLDD1156 pKa = 5.02DD1157 pKa = 4.13ATLDD1161 pKa = 3.75PATISASAGEE1171 pKa = 4.08AVLDD1175 pKa = 4.31GSTLSWTGDD1184 pKa = 3.68LAVDD1188 pKa = 3.75DD1189 pKa = 4.91VVTITYY1195 pKa = 8.13TVVVVEE1201 pKa = 4.96DD1202 pKa = 4.45GSDD1205 pKa = 3.39EE1206 pKa = 4.16LVNPVTSEE1214 pKa = 4.12DD1215 pKa = 3.6PRR1217 pKa = 11.84GTCDD1221 pKa = 3.6EE1222 pKa = 4.28EE1223 pKa = 4.78VGCEE1227 pKa = 3.88TTHH1230 pKa = 6.91LKK1232 pKa = 10.71GHH1234 pKa = 5.74FTYY1237 pKa = 10.91SKK1239 pKa = 9.45TADD1242 pKa = 3.87PGSGATVAEE1251 pKa = 4.44GDD1253 pKa = 3.96VITYY1257 pKa = 7.47TVTVSHH1263 pKa = 6.27QGKK1266 pKa = 10.09APLEE1270 pKa = 4.48DD1271 pKa = 3.25ATVVDD1276 pKa = 5.22DD1277 pKa = 4.3LTDD1280 pKa = 4.07VIDD1283 pKa = 4.01DD1284 pKa = 5.24AIYY1287 pKa = 11.0NGDD1290 pKa = 3.52VEE1292 pKa = 4.47ASSGAVRR1299 pKa = 11.84YY1300 pKa = 9.76ADD1302 pKa = 5.38SKK1304 pKa = 10.16LTWTGDD1310 pKa = 3.61LEE1312 pKa = 4.52VGQVVTFTYY1321 pKa = 9.55TVEE1324 pKa = 4.17VTGDD1328 pKa = 3.33GSDD1331 pKa = 3.42NLVNPVTTDD1340 pKa = 3.57DD1341 pKa = 4.03PRR1343 pKa = 11.84GTCDD1347 pKa = 3.61EE1348 pKa = 4.38AVGCQTEE1355 pKa = 4.72HH1356 pKa = 7.05PLEE1359 pKa = 4.68DD1360 pKa = 4.02APVPAPGLGGGGTGVGGNLPNTGAPAVLGTLLAGLLMVGAGLTVVARR1407 pKa = 11.84RR1408 pKa = 11.84RR1409 pKa = 11.84RR1410 pKa = 11.84ISLGKK1415 pKa = 10.25GQTSSLL1421 pKa = 3.73
Molecular weight: 150.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6A9UTD7|A0A6A9UTD7_9ACTN NAD(P)H-binding protein OS=Auraticoccus cholistanensis OX=2656650 GN=GC722_07720 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84THH17 pKa = 6.18GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.87GRR40 pKa = 11.84AQLSSS45 pKa = 3.28
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84THH17 pKa = 6.18GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.87GRR40 pKa = 11.84AQLSSS45 pKa = 3.28
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1128129 |
28 |
2244 |
336.0 |
35.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.112 ± 0.049 | 0.724 ± 0.01 |
5.992 ± 0.038 | 5.908 ± 0.039 |
2.597 ± 0.023 | 9.372 ± 0.039 |
2.161 ± 0.023 | 2.618 ± 0.029 |
1.261 ± 0.022 | 10.845 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.658 ± 0.018 | 1.521 ± 0.023 |
6.193 ± 0.04 | 3.132 ± 0.025 |
8.337 ± 0.05 | 5.19 ± 0.024 |
5.994 ± 0.036 | 9.871 ± 0.038 |
1.658 ± 0.019 | 1.853 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
