
Microterricola viridarii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microterricola
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
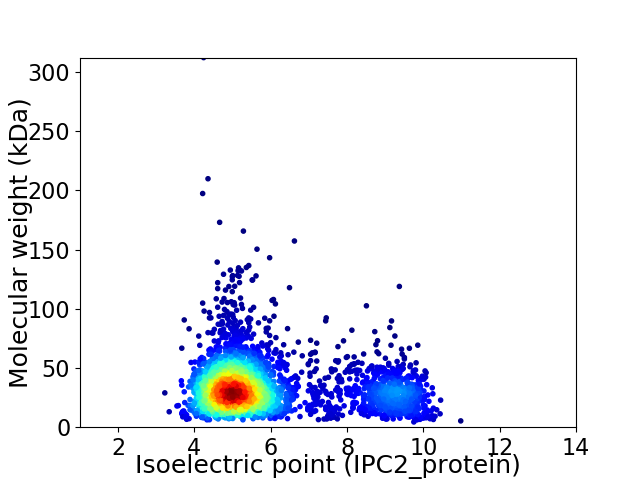
Virtual 2D-PAGE plot for 3119 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
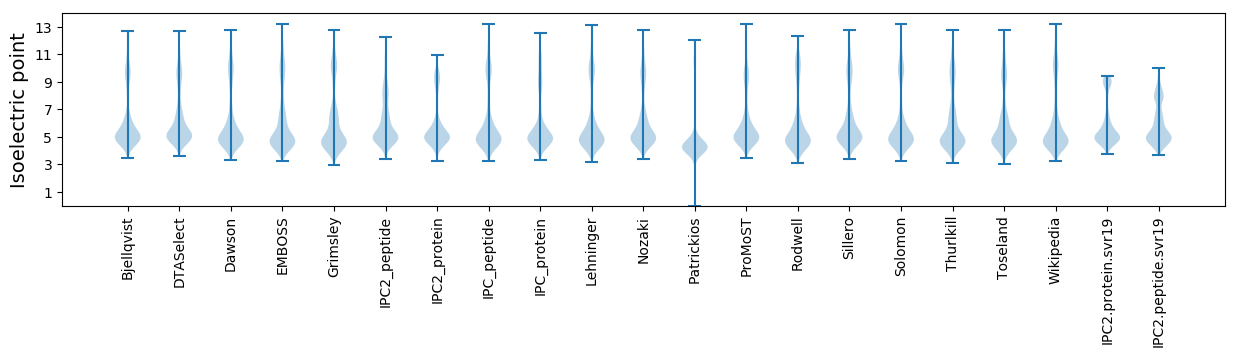
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Y0Q264|A0A0Y0Q264_9MICO HTH tetR-type domain-containing protein OS=Microterricola viridarii OX=412690 GN=AWU67_16415 PE=4 SV=1
MM1 pKa = 8.0PLTMLPTTRR10 pKa = 11.84GAARR14 pKa = 11.84VSLAALLAVGVLVGASGCTAEE35 pKa = 4.77QPASGEE41 pKa = 4.16KK42 pKa = 10.78GSWTVLTYY50 pKa = 10.76AIADD54 pKa = 3.93TDD56 pKa = 3.99LEE58 pKa = 4.58PYY60 pKa = 10.06MMEE63 pKa = 5.03DD64 pKa = 3.41LDD66 pKa = 3.99EE67 pKa = 5.66LGEE70 pKa = 4.27VGSSDD75 pKa = 3.91ALNLVALVDD84 pKa = 3.75RR85 pKa = 11.84AEE87 pKa = 5.05DD88 pKa = 3.75YY89 pKa = 11.73GDD91 pKa = 4.3DD92 pKa = 3.98PVLGMEE98 pKa = 4.76GWSGARR104 pKa = 11.84LLEE107 pKa = 4.04VGKK110 pKa = 10.76RR111 pKa = 11.84SATVLEE117 pKa = 4.18EE118 pKa = 4.05MGDD121 pKa = 3.84VNTGDD126 pKa = 3.69PAVLAEE132 pKa = 4.46FIEE135 pKa = 4.61RR136 pKa = 11.84GIAEE140 pKa = 4.37YY141 pKa = 9.15PADD144 pKa = 4.18NYY146 pKa = 11.78ALLISDD152 pKa = 5.39HH153 pKa = 5.99GASWPGVGGDD163 pKa = 3.78EE164 pKa = 4.66SAHH167 pKa = 6.57EE168 pKa = 4.27DD169 pKa = 3.67TLTLAEE175 pKa = 4.72LDD177 pKa = 3.67QAIGDD182 pKa = 4.11GLSAAGVDD190 pKa = 4.07SLDD193 pKa = 3.65LLGFDD198 pKa = 4.12ACLMATYY205 pKa = 9.83EE206 pKa = 4.35VASALAPHH214 pKa = 6.99AQRR217 pKa = 11.84LLASQEE223 pKa = 4.13LEE225 pKa = 4.43PGHH228 pKa = 6.05GWDD231 pKa = 3.72YY232 pKa = 10.15TALGILTDD240 pKa = 3.95GGVSVDD246 pKa = 3.76EE247 pKa = 4.9LGVAIIDD254 pKa = 3.86GFEE257 pKa = 3.99AQAQAEE263 pKa = 4.46GTEE266 pKa = 4.3TEE268 pKa = 4.09ITLSLVDD275 pKa = 4.08LTQMPKK281 pKa = 9.96VDD283 pKa = 3.42AALTNFTSVLTARR296 pKa = 11.84AAIAGPVVGRR306 pKa = 11.84TLAKK310 pKa = 9.11TLGFGRR316 pKa = 11.84SPDD319 pKa = 3.66PTEE322 pKa = 4.57DD323 pKa = 2.84SHH325 pKa = 6.99MADD328 pKa = 3.67LGILAGEE335 pKa = 4.25IGVDD339 pKa = 3.17ALYY342 pKa = 10.95ASDD345 pKa = 4.59AADD348 pKa = 3.7DD349 pKa = 4.02LVRR352 pKa = 11.84AINDD356 pKa = 3.32AVVYY360 pKa = 10.4KK361 pKa = 10.73VDD363 pKa = 3.58GQATRR368 pKa = 11.84GATGLSIYY376 pKa = 9.86FPPTPEE382 pKa = 4.46LFLDD386 pKa = 4.5EE387 pKa = 4.25YY388 pKa = 11.33RR389 pKa = 11.84EE390 pKa = 4.12LGNTGGWADD399 pKa = 4.09FLLAFYY405 pKa = 10.53GAGTDD410 pKa = 3.89LPVDD414 pKa = 3.26QAVRR418 pKa = 11.84FNGGAAGTLFDD429 pKa = 4.89EE430 pKa = 6.09DD431 pKa = 4.41GLTIQGTVDD440 pKa = 3.09AALIDD445 pKa = 5.28AIADD449 pKa = 3.38SYY451 pKa = 10.95IRR453 pKa = 11.84YY454 pKa = 9.83GIVEE458 pKa = 4.61DD459 pKa = 4.99DD460 pKa = 4.27GSVSYY465 pKa = 10.56IGQEE469 pKa = 3.72TATALGGDD477 pKa = 3.82SGVVQGTYY485 pKa = 11.08DD486 pKa = 3.4LTRR489 pKa = 11.84FMISDD494 pKa = 4.11GEE496 pKa = 4.34DD497 pKa = 3.06SVPAYY502 pKa = 10.31LQLAEE507 pKa = 4.65EE508 pKa = 4.41SDD510 pKa = 4.1GVISISAPMGYY521 pKa = 9.6YY522 pKa = 10.3APHH525 pKa = 7.49DD526 pKa = 3.97IDD528 pKa = 5.32GEE530 pKa = 4.76TYY532 pKa = 10.48TDD534 pKa = 3.7ALLSLTVDD542 pKa = 3.42ADD544 pKa = 3.81SGDD547 pKa = 3.99VLSEE551 pKa = 3.94TYY553 pKa = 10.97YY554 pKa = 10.93SFKK557 pKa = 10.21PGPGAYY563 pKa = 9.42GQLSVKK569 pKa = 8.74PTGLIVPEE577 pKa = 4.06VLSVLEE583 pKa = 5.34DD584 pKa = 3.67GTEE587 pKa = 3.75QWVAPTGRR595 pKa = 11.84GVYY598 pKa = 10.22AKK600 pKa = 10.48LADD603 pKa = 3.96LGYY606 pKa = 10.88DD607 pKa = 3.92FVPLASTTKK616 pKa = 10.55LVIEE620 pKa = 4.33LWVVDD625 pKa = 4.1FAGNSDD631 pKa = 3.58MVSAQVEE638 pKa = 4.77VPP640 pKa = 3.34
MM1 pKa = 8.0PLTMLPTTRR10 pKa = 11.84GAARR14 pKa = 11.84VSLAALLAVGVLVGASGCTAEE35 pKa = 4.77QPASGEE41 pKa = 4.16KK42 pKa = 10.78GSWTVLTYY50 pKa = 10.76AIADD54 pKa = 3.93TDD56 pKa = 3.99LEE58 pKa = 4.58PYY60 pKa = 10.06MMEE63 pKa = 5.03DD64 pKa = 3.41LDD66 pKa = 3.99EE67 pKa = 5.66LGEE70 pKa = 4.27VGSSDD75 pKa = 3.91ALNLVALVDD84 pKa = 3.75RR85 pKa = 11.84AEE87 pKa = 5.05DD88 pKa = 3.75YY89 pKa = 11.73GDD91 pKa = 4.3DD92 pKa = 3.98PVLGMEE98 pKa = 4.76GWSGARR104 pKa = 11.84LLEE107 pKa = 4.04VGKK110 pKa = 10.76RR111 pKa = 11.84SATVLEE117 pKa = 4.18EE118 pKa = 4.05MGDD121 pKa = 3.84VNTGDD126 pKa = 3.69PAVLAEE132 pKa = 4.46FIEE135 pKa = 4.61RR136 pKa = 11.84GIAEE140 pKa = 4.37YY141 pKa = 9.15PADD144 pKa = 4.18NYY146 pKa = 11.78ALLISDD152 pKa = 5.39HH153 pKa = 5.99GASWPGVGGDD163 pKa = 3.78EE164 pKa = 4.66SAHH167 pKa = 6.57EE168 pKa = 4.27DD169 pKa = 3.67TLTLAEE175 pKa = 4.72LDD177 pKa = 3.67QAIGDD182 pKa = 4.11GLSAAGVDD190 pKa = 4.07SLDD193 pKa = 3.65LLGFDD198 pKa = 4.12ACLMATYY205 pKa = 9.83EE206 pKa = 4.35VASALAPHH214 pKa = 6.99AQRR217 pKa = 11.84LLASQEE223 pKa = 4.13LEE225 pKa = 4.43PGHH228 pKa = 6.05GWDD231 pKa = 3.72YY232 pKa = 10.15TALGILTDD240 pKa = 3.95GGVSVDD246 pKa = 3.76EE247 pKa = 4.9LGVAIIDD254 pKa = 3.86GFEE257 pKa = 3.99AQAQAEE263 pKa = 4.46GTEE266 pKa = 4.3TEE268 pKa = 4.09ITLSLVDD275 pKa = 4.08LTQMPKK281 pKa = 9.96VDD283 pKa = 3.42AALTNFTSVLTARR296 pKa = 11.84AAIAGPVVGRR306 pKa = 11.84TLAKK310 pKa = 9.11TLGFGRR316 pKa = 11.84SPDD319 pKa = 3.66PTEE322 pKa = 4.57DD323 pKa = 2.84SHH325 pKa = 6.99MADD328 pKa = 3.67LGILAGEE335 pKa = 4.25IGVDD339 pKa = 3.17ALYY342 pKa = 10.95ASDD345 pKa = 4.59AADD348 pKa = 3.7DD349 pKa = 4.02LVRR352 pKa = 11.84AINDD356 pKa = 3.32AVVYY360 pKa = 10.4KK361 pKa = 10.73VDD363 pKa = 3.58GQATRR368 pKa = 11.84GATGLSIYY376 pKa = 9.86FPPTPEE382 pKa = 4.46LFLDD386 pKa = 4.5EE387 pKa = 4.25YY388 pKa = 11.33RR389 pKa = 11.84EE390 pKa = 4.12LGNTGGWADD399 pKa = 4.09FLLAFYY405 pKa = 10.53GAGTDD410 pKa = 3.89LPVDD414 pKa = 3.26QAVRR418 pKa = 11.84FNGGAAGTLFDD429 pKa = 4.89EE430 pKa = 6.09DD431 pKa = 4.41GLTIQGTVDD440 pKa = 3.09AALIDD445 pKa = 5.28AIADD449 pKa = 3.38SYY451 pKa = 10.95IRR453 pKa = 11.84YY454 pKa = 9.83GIVEE458 pKa = 4.61DD459 pKa = 4.99DD460 pKa = 4.27GSVSYY465 pKa = 10.56IGQEE469 pKa = 3.72TATALGGDD477 pKa = 3.82SGVVQGTYY485 pKa = 11.08DD486 pKa = 3.4LTRR489 pKa = 11.84FMISDD494 pKa = 4.11GEE496 pKa = 4.34DD497 pKa = 3.06SVPAYY502 pKa = 10.31LQLAEE507 pKa = 4.65EE508 pKa = 4.41SDD510 pKa = 4.1GVISISAPMGYY521 pKa = 9.6YY522 pKa = 10.3APHH525 pKa = 7.49DD526 pKa = 3.97IDD528 pKa = 5.32GEE530 pKa = 4.76TYY532 pKa = 10.48TDD534 pKa = 3.7ALLSLTVDD542 pKa = 3.42ADD544 pKa = 3.81SGDD547 pKa = 3.99VLSEE551 pKa = 3.94TYY553 pKa = 10.97YY554 pKa = 10.93SFKK557 pKa = 10.21PGPGAYY563 pKa = 9.42GQLSVKK569 pKa = 8.74PTGLIVPEE577 pKa = 4.06VLSVLEE583 pKa = 5.34DD584 pKa = 3.67GTEE587 pKa = 3.75QWVAPTGRR595 pKa = 11.84GVYY598 pKa = 10.22AKK600 pKa = 10.48LADD603 pKa = 3.96LGYY606 pKa = 10.88DD607 pKa = 3.92FVPLASTTKK616 pKa = 10.55LVIEE620 pKa = 4.33LWVVDD625 pKa = 4.1FAGNSDD631 pKa = 3.58MVSAQVEE638 pKa = 4.77VPP640 pKa = 3.34
Molecular weight: 66.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A120I1A9|A0A120I1A9_9MICO Uncharacterized protein OS=Microterricola viridarii OX=412690 GN=AWU67_15615 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILGARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.21GRR40 pKa = 11.84VEE42 pKa = 4.17LSAA45 pKa = 5.19
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILGARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.21GRR40 pKa = 11.84VEE42 pKa = 4.17LSAA45 pKa = 5.19
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1022284 |
38 |
3052 |
327.8 |
34.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.975 ± 0.062 | 0.496 ± 0.011 |
5.59 ± 0.034 | 5.565 ± 0.045 |
3.277 ± 0.026 | 9.097 ± 0.042 |
1.918 ± 0.02 | 4.739 ± 0.033 |
2.117 ± 0.034 | 10.523 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.868 ± 0.019 | 2.265 ± 0.025 |
5.26 ± 0.033 | 2.973 ± 0.022 |
6.463 ± 0.044 | 5.916 ± 0.031 |
5.953 ± 0.047 | 8.61 ± 0.041 |
1.437 ± 0.02 | 1.961 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
