
Flavobacterium sp. AG291
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; unclassified Flavobacterium
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
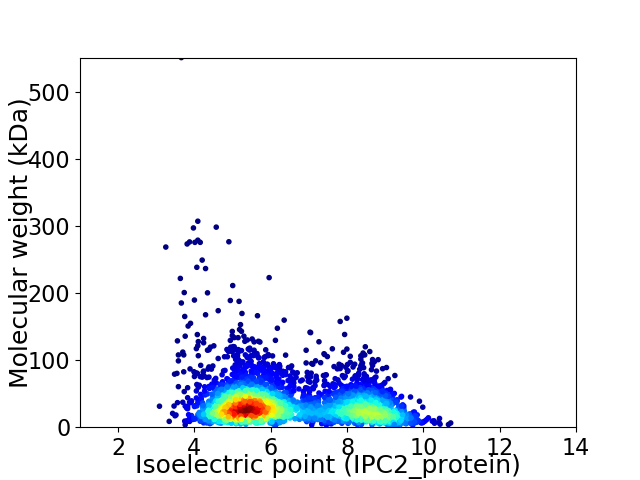
Virtual 2D-PAGE plot for 3455 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A370EB97|A0A370EB97_9FLAO Alanine dehydrogenase OS=Flavobacterium sp. AG291 OX=2184000 GN=DEU42_12017 PE=3 SV=1
MM1 pKa = 7.52KK2 pKa = 10.46RR3 pKa = 11.84LLPVLLILLGVYY15 pKa = 7.59TANAQVGIGTTTPNPSAQLQIVSGDD40 pKa = 3.38KK41 pKa = 10.88GVLIPQVQLTSTTDD55 pKa = 2.86TTTITNGNVQSLLVYY70 pKa = 10.68NITSNSQIKK79 pKa = 8.82PGYY82 pKa = 8.67YY83 pKa = 8.62YY84 pKa = 9.93WYY86 pKa = 7.96VNKK89 pKa = 9.74WYY91 pKa = 10.56RR92 pKa = 11.84LLSDD96 pKa = 4.02GDD98 pKa = 3.8AGQGDD103 pKa = 4.49SAQPIIDD110 pKa = 4.43NGNGTYY116 pKa = 9.96TYY118 pKa = 8.93TNPDD122 pKa = 3.06GSVTTIDD129 pKa = 3.45VPGNQTLTFLVYY141 pKa = 10.88NPTAKK146 pKa = 9.89TLTYY150 pKa = 9.41TAEE153 pKa = 3.96NGFNTVIDD161 pKa = 4.24LDD163 pKa = 4.23AVVKK167 pKa = 10.79ASEE170 pKa = 4.37TVTTLVDD177 pKa = 3.48NGNGTVTFTNEE188 pKa = 3.45AGVATTINLAGGTQGVGIASTVDD211 pKa = 3.11NGDD214 pKa = 3.19GTFTINYY221 pKa = 7.68TDD223 pKa = 3.52GTSFTTSNLTGPQGPQGQAGPAAAPGVGIASTVDD257 pKa = 3.11NGDD260 pKa = 3.19GTFTINYY267 pKa = 7.15TNGTSFTTTSLLGPQGPVGATGAQGVAGATGADD300 pKa = 3.4GKK302 pKa = 11.16GITSTTDD309 pKa = 2.77NGNGTFTINYY319 pKa = 7.65TDD321 pKa = 3.52GTSFTTSNLTGATGAQGATGADD343 pKa = 3.15GKK345 pKa = 11.19GIVSTTDD352 pKa = 2.83NGNGTFTINFTDD364 pKa = 3.97GTSFTTSNLTGPQGATGTQGIQGPQGATGNDD395 pKa = 3.83GVAGPQGATGADD407 pKa = 3.26GKK409 pKa = 11.34GIATTVDD416 pKa = 3.09NGNGTFTINYY426 pKa = 7.65TDD428 pKa = 3.52GTSFTTSNLTGPQGATGTQGIQGPQGATGNDD459 pKa = 3.83GVAGPQGATGADD471 pKa = 3.29GKK473 pKa = 11.14GIASTTDD480 pKa = 2.91NGNGTFTINYY490 pKa = 7.65TDD492 pKa = 3.52GTSFTTSNLTGPQGATGTQGIQGPQGATGNDD523 pKa = 3.83GVAGPQGATGADD535 pKa = 3.29GKK537 pKa = 11.19GIASTVDD544 pKa = 3.01NGNGTFTINYY554 pKa = 7.65TDD556 pKa = 3.52GTSFTTSNLTGPQGATGTQGIQGPQGATGNDD587 pKa = 3.83GVAGPQGATGADD599 pKa = 3.29GKK601 pKa = 11.14GIASTTDD608 pKa = 2.91NGNGTFTT615 pKa = 4.11
MM1 pKa = 7.52KK2 pKa = 10.46RR3 pKa = 11.84LLPVLLILLGVYY15 pKa = 7.59TANAQVGIGTTTPNPSAQLQIVSGDD40 pKa = 3.38KK41 pKa = 10.88GVLIPQVQLTSTTDD55 pKa = 2.86TTTITNGNVQSLLVYY70 pKa = 10.68NITSNSQIKK79 pKa = 8.82PGYY82 pKa = 8.67YY83 pKa = 8.62YY84 pKa = 9.93WYY86 pKa = 7.96VNKK89 pKa = 9.74WYY91 pKa = 10.56RR92 pKa = 11.84LLSDD96 pKa = 4.02GDD98 pKa = 3.8AGQGDD103 pKa = 4.49SAQPIIDD110 pKa = 4.43NGNGTYY116 pKa = 9.96TYY118 pKa = 8.93TNPDD122 pKa = 3.06GSVTTIDD129 pKa = 3.45VPGNQTLTFLVYY141 pKa = 10.88NPTAKK146 pKa = 9.89TLTYY150 pKa = 9.41TAEE153 pKa = 3.96NGFNTVIDD161 pKa = 4.24LDD163 pKa = 4.23AVVKK167 pKa = 10.79ASEE170 pKa = 4.37TVTTLVDD177 pKa = 3.48NGNGTVTFTNEE188 pKa = 3.45AGVATTINLAGGTQGVGIASTVDD211 pKa = 3.11NGDD214 pKa = 3.19GTFTINYY221 pKa = 7.68TDD223 pKa = 3.52GTSFTTSNLTGPQGPQGQAGPAAAPGVGIASTVDD257 pKa = 3.11NGDD260 pKa = 3.19GTFTINYY267 pKa = 7.15TNGTSFTTTSLLGPQGPVGATGAQGVAGATGADD300 pKa = 3.4GKK302 pKa = 11.16GITSTTDD309 pKa = 2.77NGNGTFTINYY319 pKa = 7.65TDD321 pKa = 3.52GTSFTTSNLTGATGAQGATGADD343 pKa = 3.15GKK345 pKa = 11.19GIVSTTDD352 pKa = 2.83NGNGTFTINFTDD364 pKa = 3.97GTSFTTSNLTGPQGATGTQGIQGPQGATGNDD395 pKa = 3.83GVAGPQGATGADD407 pKa = 3.26GKK409 pKa = 11.34GIATTVDD416 pKa = 3.09NGNGTFTINYY426 pKa = 7.65TDD428 pKa = 3.52GTSFTTSNLTGPQGATGTQGIQGPQGATGNDD459 pKa = 3.83GVAGPQGATGADD471 pKa = 3.29GKK473 pKa = 11.14GIASTTDD480 pKa = 2.91NGNGTFTINYY490 pKa = 7.65TDD492 pKa = 3.52GTSFTTSNLTGPQGATGTQGIQGPQGATGNDD523 pKa = 3.83GVAGPQGATGADD535 pKa = 3.29GKK537 pKa = 11.19GIASTVDD544 pKa = 3.01NGNGTFTINYY554 pKa = 7.65TDD556 pKa = 3.52GTSFTTSNLTGPQGATGTQGIQGPQGATGNDD587 pKa = 3.83GVAGPQGATGADD599 pKa = 3.29GKK601 pKa = 11.14GIASTTDD608 pKa = 2.91NGNGTFTT615 pKa = 4.11
Molecular weight: 60.38 kDa
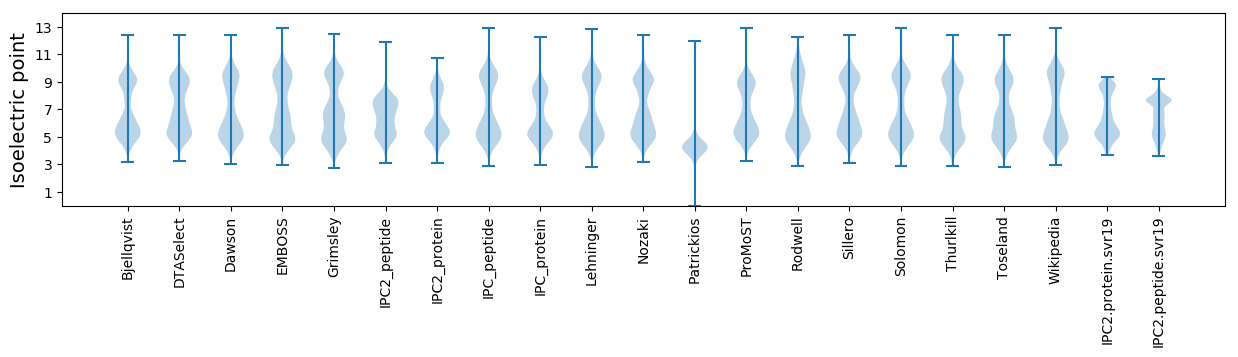
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A370EM15|A0A370EM15_9FLAO Uncharacterized protein OS=Flavobacterium sp. AG291 OX=2184000 GN=DEU42_10668 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.53HH17 pKa = 4.24GFMEE21 pKa = 4.81RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.53HH17 pKa = 4.24GFMEE21 pKa = 4.81RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1150073 |
26 |
5333 |
332.9 |
37.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.17 ± 0.054 | 0.816 ± 0.018 |
5.501 ± 0.034 | 6.238 ± 0.053 |
4.981 ± 0.041 | 6.724 ± 0.047 |
1.694 ± 0.023 | 7.441 ± 0.05 |
7.243 ± 0.078 | 8.809 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.322 ± 0.025 | 6.06 ± 0.039 |
3.647 ± 0.033 | 3.336 ± 0.024 |
3.32 ± 0.035 | 6.309 ± 0.04 |
6.549 ± 0.106 | 6.524 ± 0.043 |
1.053 ± 0.016 | 4.265 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
