
Microviridae sp. ctQch27
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 9.09
Get precalculated fractions of proteins
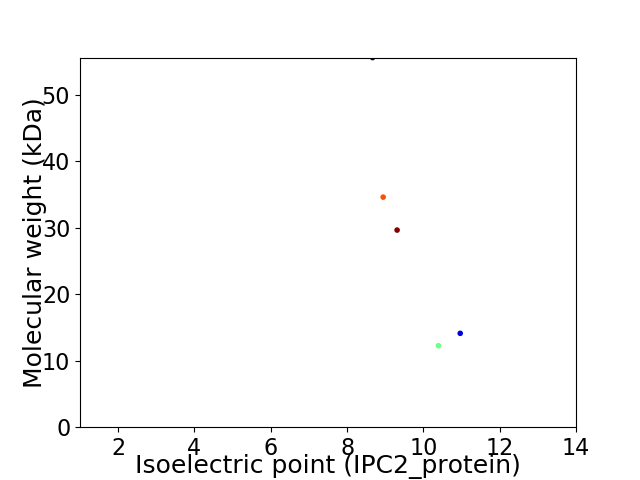
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W8W3|A0A5Q2W8W3_9VIRU Major capsid protein OS=Microviridae sp. ctQch27 OX=2656699 PE=3 SV=1
MM1 pKa = 7.08KK2 pKa = 10.24RR3 pKa = 11.84SVFSLSHH10 pKa = 6.36HH11 pKa = 6.51KK12 pKa = 10.53LLSFDD17 pKa = 3.38MGEE20 pKa = 4.12LVPIGMTEE28 pKa = 4.11VLPGDD33 pKa = 4.0TMQQATTMLVRR44 pKa = 11.84ASPLLAPVMHH54 pKa = 6.74PVDD57 pKa = 3.67VRR59 pKa = 11.84IHH61 pKa = 5.6HH62 pKa = 6.51WFVPHH67 pKa = 6.45RR68 pKa = 11.84LVWADD73 pKa = 3.33FEE75 pKa = 4.65KK76 pKa = 10.68FITGGPDD83 pKa = 3.06GLDD86 pKa = 2.67ASVFPTITFGGAIPAGTVMDD106 pKa = 4.4YY107 pKa = 11.41LGMPLGVAGLTVSALPFRR125 pKa = 11.84AYY127 pKa = 11.13NLIWNNWYY135 pKa = 10.0RR136 pKa = 11.84DD137 pKa = 3.45QDD139 pKa = 4.16LEE141 pKa = 4.17TALAVSTASGNDD153 pKa = 2.95VTTNTNLKK161 pKa = 8.83SCSWEE166 pKa = 3.51KK167 pKa = 11.19DD168 pKa = 3.38YY169 pKa = 9.31FTSARR174 pKa = 11.84PWEE177 pKa = 4.43QKK179 pKa = 10.73GPTVTVPLGTTAPVMGLGFANTPSQMAASTVMRR212 pKa = 11.84QTTPVANEE220 pKa = 4.22TVNNPPWWMITNAANTMGVKK240 pKa = 10.03ADD242 pKa = 3.7SAASGTTKK250 pKa = 10.39PSVFADD256 pKa = 3.94LTNASAVTVNTLRR269 pKa = 11.84QALAFQRR276 pKa = 11.84YY277 pKa = 7.43EE278 pKa = 3.67EE279 pKa = 3.99ARR281 pKa = 11.84ARR283 pKa = 11.84FGSRR287 pKa = 11.84YY288 pKa = 9.26IEE290 pKa = 3.9YY291 pKa = 10.5LRR293 pKa = 11.84YY294 pKa = 10.12LGVKK298 pKa = 10.15SSDD301 pKa = 3.04GRR303 pKa = 11.84LQVPEE308 pKa = 4.01YY309 pKa = 10.71LGGGRR314 pKa = 11.84DD315 pKa = 3.59TIQFSEE321 pKa = 4.24ILQTAADD328 pKa = 3.88GTNPVGTMRR337 pKa = 11.84GHH339 pKa = 7.34GIAGMRR345 pKa = 11.84SNRR348 pKa = 11.84YY349 pKa = 8.12RR350 pKa = 11.84RR351 pKa = 11.84FFEE354 pKa = 3.53EE355 pKa = 3.29HH356 pKa = 6.73GYY358 pKa = 10.71VMSLASVRR366 pKa = 11.84PKK368 pKa = 9.96TMYY371 pKa = 9.4MQGLARR377 pKa = 11.84TWNRR381 pKa = 11.84RR382 pKa = 11.84VKK384 pKa = 10.55EE385 pKa = 4.12DD386 pKa = 3.37FWQKK390 pKa = 9.59EE391 pKa = 4.02LQFIGQQAIKK401 pKa = 10.73NKK403 pKa = 9.39EE404 pKa = 3.82LYY406 pKa = 9.63AAHH409 pKa = 7.36ASPDD413 pKa = 3.51GTFGYY418 pKa = 9.77QDD420 pKa = 3.56RR421 pKa = 11.84YY422 pKa = 10.84DD423 pKa = 3.41EE424 pKa = 4.2YY425 pKa = 11.03RR426 pKa = 11.84RR427 pKa = 11.84SEE429 pKa = 4.09SSVSGEE435 pKa = 3.49FRR437 pKa = 11.84TVLNFWHH444 pKa = 7.24FARR447 pKa = 11.84EE448 pKa = 4.28FGSSPALNGTFVSCVPTEE466 pKa = 3.8RR467 pKa = 11.84TFAVSTNDD475 pKa = 3.5VLWVMAKK482 pKa = 10.28HH483 pKa = 6.63SIQARR488 pKa = 11.84RR489 pKa = 11.84LVAKK493 pKa = 9.13TGRR496 pKa = 11.84SMTFF500 pKa = 2.75
MM1 pKa = 7.08KK2 pKa = 10.24RR3 pKa = 11.84SVFSLSHH10 pKa = 6.36HH11 pKa = 6.51KK12 pKa = 10.53LLSFDD17 pKa = 3.38MGEE20 pKa = 4.12LVPIGMTEE28 pKa = 4.11VLPGDD33 pKa = 4.0TMQQATTMLVRR44 pKa = 11.84ASPLLAPVMHH54 pKa = 6.74PVDD57 pKa = 3.67VRR59 pKa = 11.84IHH61 pKa = 5.6HH62 pKa = 6.51WFVPHH67 pKa = 6.45RR68 pKa = 11.84LVWADD73 pKa = 3.33FEE75 pKa = 4.65KK76 pKa = 10.68FITGGPDD83 pKa = 3.06GLDD86 pKa = 2.67ASVFPTITFGGAIPAGTVMDD106 pKa = 4.4YY107 pKa = 11.41LGMPLGVAGLTVSALPFRR125 pKa = 11.84AYY127 pKa = 11.13NLIWNNWYY135 pKa = 10.0RR136 pKa = 11.84DD137 pKa = 3.45QDD139 pKa = 4.16LEE141 pKa = 4.17TALAVSTASGNDD153 pKa = 2.95VTTNTNLKK161 pKa = 8.83SCSWEE166 pKa = 3.51KK167 pKa = 11.19DD168 pKa = 3.38YY169 pKa = 9.31FTSARR174 pKa = 11.84PWEE177 pKa = 4.43QKK179 pKa = 10.73GPTVTVPLGTTAPVMGLGFANTPSQMAASTVMRR212 pKa = 11.84QTTPVANEE220 pKa = 4.22TVNNPPWWMITNAANTMGVKK240 pKa = 10.03ADD242 pKa = 3.7SAASGTTKK250 pKa = 10.39PSVFADD256 pKa = 3.94LTNASAVTVNTLRR269 pKa = 11.84QALAFQRR276 pKa = 11.84YY277 pKa = 7.43EE278 pKa = 3.67EE279 pKa = 3.99ARR281 pKa = 11.84ARR283 pKa = 11.84FGSRR287 pKa = 11.84YY288 pKa = 9.26IEE290 pKa = 3.9YY291 pKa = 10.5LRR293 pKa = 11.84YY294 pKa = 10.12LGVKK298 pKa = 10.15SSDD301 pKa = 3.04GRR303 pKa = 11.84LQVPEE308 pKa = 4.01YY309 pKa = 10.71LGGGRR314 pKa = 11.84DD315 pKa = 3.59TIQFSEE321 pKa = 4.24ILQTAADD328 pKa = 3.88GTNPVGTMRR337 pKa = 11.84GHH339 pKa = 7.34GIAGMRR345 pKa = 11.84SNRR348 pKa = 11.84YY349 pKa = 8.12RR350 pKa = 11.84RR351 pKa = 11.84FFEE354 pKa = 3.53EE355 pKa = 3.29HH356 pKa = 6.73GYY358 pKa = 10.71VMSLASVRR366 pKa = 11.84PKK368 pKa = 9.96TMYY371 pKa = 9.4MQGLARR377 pKa = 11.84TWNRR381 pKa = 11.84RR382 pKa = 11.84VKK384 pKa = 10.55EE385 pKa = 4.12DD386 pKa = 3.37FWQKK390 pKa = 9.59EE391 pKa = 4.02LQFIGQQAIKK401 pKa = 10.73NKK403 pKa = 9.39EE404 pKa = 3.82LYY406 pKa = 9.63AAHH409 pKa = 7.36ASPDD413 pKa = 3.51GTFGYY418 pKa = 9.77QDD420 pKa = 3.56RR421 pKa = 11.84YY422 pKa = 10.84DD423 pKa = 3.41EE424 pKa = 4.2YY425 pKa = 11.03RR426 pKa = 11.84RR427 pKa = 11.84SEE429 pKa = 4.09SSVSGEE435 pKa = 3.49FRR437 pKa = 11.84TVLNFWHH444 pKa = 7.24FARR447 pKa = 11.84EE448 pKa = 4.28FGSSPALNGTFVSCVPTEE466 pKa = 3.8RR467 pKa = 11.84TFAVSTNDD475 pKa = 3.5VLWVMAKK482 pKa = 10.28HH483 pKa = 6.63SIQARR488 pKa = 11.84RR489 pKa = 11.84LVAKK493 pKa = 9.13TGRR496 pKa = 11.84SMTFF500 pKa = 2.75
Molecular weight: 55.58 kDa
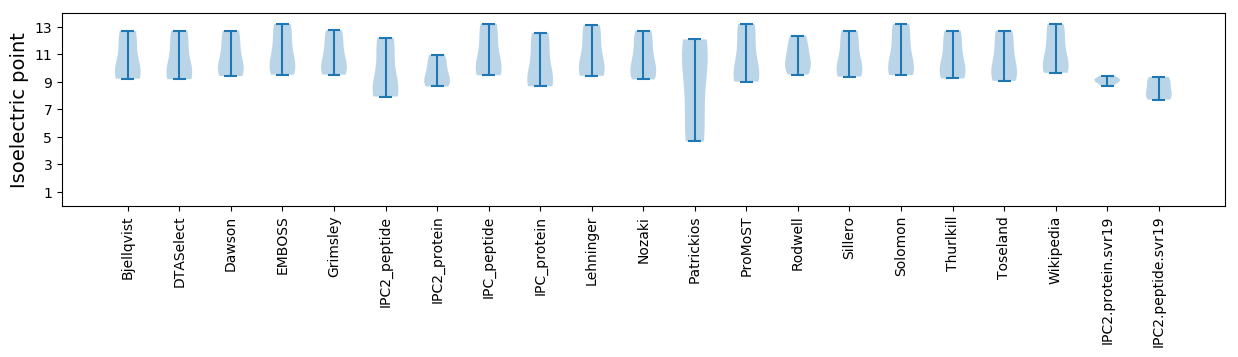
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W671|A0A5Q2W671_9VIRU Putative minor capsid protein OS=Microviridae sp. ctQch27 OX=2656699 PE=4 SV=1
MM1 pKa = 7.18SLKK4 pKa = 10.38RR5 pKa = 11.84LRR7 pKa = 11.84KK8 pKa = 8.93RR9 pKa = 11.84RR10 pKa = 11.84ISRR13 pKa = 11.84LVRR16 pKa = 11.84TRR18 pKa = 11.84VWRR21 pKa = 11.84IGSLTSILLRR31 pKa = 11.84LRR33 pKa = 11.84STFAGRR39 pKa = 11.84KK40 pKa = 7.14PASLCMRR47 pKa = 11.84KK48 pKa = 9.66SSLMKK53 pKa = 10.01KK54 pKa = 10.12VSWFLLKK61 pKa = 10.86GILKK65 pKa = 9.68AFPRR69 pKa = 11.84SRR71 pKa = 11.84PRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84LLDD78 pKa = 3.63GPRR81 pKa = 11.84SPSRR85 pKa = 11.84AAVGRR90 pKa = 11.84SSRR93 pKa = 11.84PLTGFRR99 pKa = 11.84ARR101 pKa = 11.84LFLRR105 pKa = 11.84LLGGVVGNAVHH116 pKa = 7.05SPGEE120 pKa = 3.96LKK122 pKa = 10.81ARR124 pKa = 3.95
MM1 pKa = 7.18SLKK4 pKa = 10.38RR5 pKa = 11.84LRR7 pKa = 11.84KK8 pKa = 8.93RR9 pKa = 11.84RR10 pKa = 11.84ISRR13 pKa = 11.84LVRR16 pKa = 11.84TRR18 pKa = 11.84VWRR21 pKa = 11.84IGSLTSILLRR31 pKa = 11.84LRR33 pKa = 11.84STFAGRR39 pKa = 11.84KK40 pKa = 7.14PASLCMRR47 pKa = 11.84KK48 pKa = 9.66SSLMKK53 pKa = 10.01KK54 pKa = 10.12VSWFLLKK61 pKa = 10.86GILKK65 pKa = 9.68AFPRR69 pKa = 11.84SRR71 pKa = 11.84PRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84LLDD78 pKa = 3.63GPRR81 pKa = 11.84SPSRR85 pKa = 11.84AAVGRR90 pKa = 11.84SSRR93 pKa = 11.84PLTGFRR99 pKa = 11.84ARR101 pKa = 11.84LFLRR105 pKa = 11.84LLGGVVGNAVHH116 pKa = 7.05SPGEE120 pKa = 3.96LKK122 pKa = 10.81ARR124 pKa = 3.95
Molecular weight: 14.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1303 |
107 |
500 |
260.6 |
29.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.44 ± 1.1 | 1.074 ± 0.538 |
3.147 ± 0.536 | 4.988 ± 0.854 |
4.068 ± 0.601 | 8.289 ± 0.609 |
1.765 ± 0.301 | 2.993 ± 0.269 |
4.451 ± 0.62 | 8.365 ± 0.934 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.3 ± 0.31 | 2.916 ± 0.791 |
5.602 ± 0.655 | 3.761 ± 0.707 |
10.207 ± 2.234 | 7.368 ± 0.57 |
5.526 ± 1.542 | 7.368 ± 1.085 |
2.686 ± 0.45 | 2.686 ± 0.413 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
