
Wenling narna-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.67
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
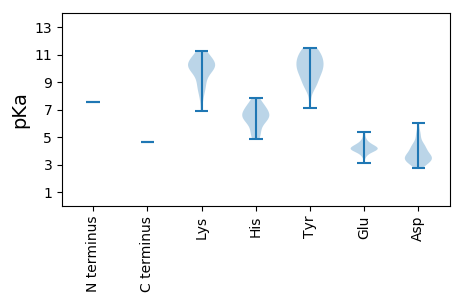
>tr|A0A1L3KIM5|A0A1L3KIM5_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 2 OX=1923502 PE=4 SV=1
MM1 pKa = 7.58SPRR4 pKa = 11.84NLFPSYY10 pKa = 11.42GDD12 pKa = 3.34GCSSVNSTEE21 pKa = 3.85NTLASILEE29 pKa = 4.65SPFHH33 pKa = 6.14PAWSAVVEE41 pKa = 4.36GFFGNEE47 pKa = 3.95VVTDD51 pKa = 3.47THH53 pKa = 7.85VDD55 pKa = 3.26EE56 pKa = 4.57LAIYY60 pKa = 10.4EE61 pKa = 4.8DD62 pKa = 4.39GLDD65 pKa = 3.47LSAYY69 pKa = 7.12STSSVEE75 pKa = 4.01QVVRR79 pKa = 11.84CCEE82 pKa = 4.2VYY84 pKa = 10.92SFIYY88 pKa = 9.81DD89 pKa = 3.18QYY91 pKa = 10.67GYY93 pKa = 11.09KK94 pKa = 10.37FLFSPTDD101 pKa = 3.15QRR103 pKa = 11.84LWLEE107 pKa = 4.67LGDD110 pKa = 4.3KK111 pKa = 10.78DD112 pKa = 5.12LLEE115 pKa = 5.36VYY117 pKa = 9.85LKK119 pKa = 10.39WITANILAAVWEE131 pKa = 4.05QDD133 pKa = 3.3EE134 pKa = 4.99LVPLPSSLLDD144 pKa = 3.94FSARR148 pKa = 11.84FPAFGLTTRR157 pKa = 11.84DD158 pKa = 3.23DD159 pKa = 3.68RR160 pKa = 11.84LFRR163 pKa = 11.84SMIRR167 pKa = 11.84SSRR170 pKa = 11.84RR171 pKa = 11.84SKK173 pKa = 9.1KK174 pKa = 6.89QQRR177 pKa = 11.84FVYY180 pKa = 9.7TLYY183 pKa = 10.45QGKK186 pKa = 9.96AGCNAMRR193 pKa = 11.84PEE195 pKa = 4.09LVAAKK200 pKa = 9.9VVSAVEE206 pKa = 4.11TLATPPEE213 pKa = 4.46GEE215 pKa = 4.06EE216 pKa = 4.15SIGILGHH223 pKa = 7.43LITKK227 pKa = 10.06EE228 pKa = 4.06DD229 pKa = 3.26IRR231 pKa = 11.84GQLLRR236 pKa = 11.84TVTEE240 pKa = 4.35IYY242 pKa = 9.18GTGHH246 pKa = 5.11ARR248 pKa = 11.84EE249 pKa = 3.96RR250 pKa = 11.84RR251 pKa = 11.84TPGSRR256 pKa = 11.84RR257 pKa = 11.84MASSKK262 pKa = 11.08SSFQYY267 pKa = 9.93TRR269 pKa = 11.84SKK271 pKa = 11.11GGAHH275 pKa = 6.73EE276 pKa = 4.31FLVDD280 pKa = 3.54NAPASMFIQPRR291 pKa = 11.84ILSGFVVYY299 pKa = 8.69KK300 pKa = 8.71TQVRR304 pKa = 11.84PYY306 pKa = 8.49YY307 pKa = 8.73TTFQAEE313 pKa = 4.31DD314 pKa = 3.39WEE316 pKa = 4.39EE317 pKa = 3.84LQEE320 pKa = 4.11LSRR323 pKa = 11.84RR324 pKa = 11.84AAWSEE329 pKa = 3.74EE330 pKa = 3.74MDD332 pKa = 4.54CYY334 pKa = 10.86PVGLPEE340 pKa = 4.1AFKK343 pKa = 11.24VRR345 pKa = 11.84VITKK349 pKa = 10.1GAVHH353 pKa = 7.27HH354 pKa = 6.45YY355 pKa = 9.81NLARR359 pKa = 11.84RR360 pKa = 11.84WQPAMWEE367 pKa = 4.08PLANHH372 pKa = 6.36PTFEE376 pKa = 4.87LVGTPNCQRR385 pKa = 11.84IMNRR389 pKa = 11.84FLAKK393 pKa = 10.09CDD395 pKa = 4.01PDD397 pKa = 3.93DD398 pKa = 5.29DD399 pKa = 4.47RR400 pKa = 11.84AFTSGDD406 pKa = 3.42YY407 pKa = 11.15ASATDD412 pKa = 4.39YY413 pKa = 11.46LDD415 pKa = 4.3SDD417 pKa = 4.42LSRR420 pKa = 11.84SCLEE424 pKa = 4.05YY425 pKa = 10.13VCNALRR431 pKa = 11.84VPFEE435 pKa = 4.14DD436 pKa = 3.52TVILAEE442 pKa = 4.33ALTDD446 pKa = 3.63HH447 pKa = 7.06RR448 pKa = 11.84LHH450 pKa = 6.47YY451 pKa = 10.4VDD453 pKa = 4.94SEE455 pKa = 4.21GVEE458 pKa = 3.9QVKK461 pKa = 7.79QQRR464 pKa = 11.84RR465 pKa = 11.84GQLMGSPISFPILCLFNAALTRR487 pKa = 11.84YY488 pKa = 8.88ALEE491 pKa = 4.16IASCDD496 pKa = 3.65SVALDD501 pKa = 5.36LEE503 pKa = 4.59DD504 pKa = 6.02LPMLINGDD512 pKa = 4.11DD513 pKa = 4.02LLCRR517 pKa = 11.84TNPLEE522 pKa = 3.87YY523 pKa = 10.49LVWKK527 pKa = 10.26DD528 pKa = 3.07IVNFGGLTPSIGKK541 pKa = 8.67NFRR544 pKa = 11.84HH545 pKa = 5.35RR546 pKa = 11.84TIGTINSEE554 pKa = 3.5MWTFKK559 pKa = 9.85HH560 pKa = 6.32TIHH563 pKa = 6.83GCPANGTDD571 pKa = 2.94TAFYY575 pKa = 9.61QGKK578 pKa = 8.26RR579 pKa = 11.84QPIIEE584 pKa = 4.39IGLVRR589 pKa = 11.84GSVKK593 pKa = 10.44RR594 pKa = 11.84GTINRR599 pKa = 11.84NVDD602 pKa = 3.49EE603 pKa = 5.25LSPFVDD609 pKa = 3.08SSKK612 pKa = 10.53FGKK615 pKa = 10.41SKK617 pKa = 8.53EE618 pKa = 4.12QCWNRR623 pKa = 11.84FLDD626 pKa = 3.67TCPNRR631 pKa = 11.84LTAYY635 pKa = 10.0DD636 pKa = 3.87FLWRR640 pKa = 11.84SVSSDD645 pKa = 2.73ILEE648 pKa = 4.23SLPRR652 pKa = 11.84GMPMCCPTWLGGAGFPLPPLGHH674 pKa = 7.28ALRR677 pKa = 11.84DD678 pKa = 3.57RR679 pKa = 11.84RR680 pKa = 11.84EE681 pKa = 3.58PSAYY685 pKa = 9.47QRR687 pKa = 11.84LLARR691 pKa = 11.84YY692 pKa = 9.27LYY694 pKa = 8.96DD695 pKa = 3.06TYY697 pKa = 11.46GYY699 pKa = 10.71GLGKK703 pKa = 10.1RR704 pKa = 11.84YY705 pKa = 10.64VSMLEE710 pKa = 4.19TGSLPANLVKK720 pKa = 10.59EE721 pKa = 4.32MNTDD725 pKa = 2.9NGIRR729 pKa = 11.84DD730 pKa = 3.68VLGVPKK736 pKa = 9.91PVPEE740 pKa = 4.51SADD743 pKa = 3.38SVDD746 pKa = 4.21GIKK749 pKa = 11.04GNLHH753 pKa = 5.6SEE755 pKa = 4.27KK756 pKa = 10.24TPLTPLPQTHH766 pKa = 6.83FFMCGLINHH775 pKa = 6.21TEE777 pKa = 4.04EE778 pKa = 4.22EE779 pKa = 4.43STGRR783 pKa = 11.84FSCEE787 pKa = 3.08KK788 pKa = 9.17MYY790 pKa = 11.24SRR792 pKa = 11.84LQHH795 pKa = 6.56KK796 pKa = 9.41ALKK799 pKa = 9.78HH800 pKa = 4.86GRR802 pKa = 11.84GPLDD806 pKa = 3.56CKK808 pKa = 10.67FFLPTPPSLKK818 pKa = 10.17LEE820 pKa = 4.13SYY822 pKa = 10.19VFQEE826 pKa = 4.43VVAA829 pKa = 4.65
MM1 pKa = 7.58SPRR4 pKa = 11.84NLFPSYY10 pKa = 11.42GDD12 pKa = 3.34GCSSVNSTEE21 pKa = 3.85NTLASILEE29 pKa = 4.65SPFHH33 pKa = 6.14PAWSAVVEE41 pKa = 4.36GFFGNEE47 pKa = 3.95VVTDD51 pKa = 3.47THH53 pKa = 7.85VDD55 pKa = 3.26EE56 pKa = 4.57LAIYY60 pKa = 10.4EE61 pKa = 4.8DD62 pKa = 4.39GLDD65 pKa = 3.47LSAYY69 pKa = 7.12STSSVEE75 pKa = 4.01QVVRR79 pKa = 11.84CCEE82 pKa = 4.2VYY84 pKa = 10.92SFIYY88 pKa = 9.81DD89 pKa = 3.18QYY91 pKa = 10.67GYY93 pKa = 11.09KK94 pKa = 10.37FLFSPTDD101 pKa = 3.15QRR103 pKa = 11.84LWLEE107 pKa = 4.67LGDD110 pKa = 4.3KK111 pKa = 10.78DD112 pKa = 5.12LLEE115 pKa = 5.36VYY117 pKa = 9.85LKK119 pKa = 10.39WITANILAAVWEE131 pKa = 4.05QDD133 pKa = 3.3EE134 pKa = 4.99LVPLPSSLLDD144 pKa = 3.94FSARR148 pKa = 11.84FPAFGLTTRR157 pKa = 11.84DD158 pKa = 3.23DD159 pKa = 3.68RR160 pKa = 11.84LFRR163 pKa = 11.84SMIRR167 pKa = 11.84SSRR170 pKa = 11.84RR171 pKa = 11.84SKK173 pKa = 9.1KK174 pKa = 6.89QQRR177 pKa = 11.84FVYY180 pKa = 9.7TLYY183 pKa = 10.45QGKK186 pKa = 9.96AGCNAMRR193 pKa = 11.84PEE195 pKa = 4.09LVAAKK200 pKa = 9.9VVSAVEE206 pKa = 4.11TLATPPEE213 pKa = 4.46GEE215 pKa = 4.06EE216 pKa = 4.15SIGILGHH223 pKa = 7.43LITKK227 pKa = 10.06EE228 pKa = 4.06DD229 pKa = 3.26IRR231 pKa = 11.84GQLLRR236 pKa = 11.84TVTEE240 pKa = 4.35IYY242 pKa = 9.18GTGHH246 pKa = 5.11ARR248 pKa = 11.84EE249 pKa = 3.96RR250 pKa = 11.84RR251 pKa = 11.84TPGSRR256 pKa = 11.84RR257 pKa = 11.84MASSKK262 pKa = 11.08SSFQYY267 pKa = 9.93TRR269 pKa = 11.84SKK271 pKa = 11.11GGAHH275 pKa = 6.73EE276 pKa = 4.31FLVDD280 pKa = 3.54NAPASMFIQPRR291 pKa = 11.84ILSGFVVYY299 pKa = 8.69KK300 pKa = 8.71TQVRR304 pKa = 11.84PYY306 pKa = 8.49YY307 pKa = 8.73TTFQAEE313 pKa = 4.31DD314 pKa = 3.39WEE316 pKa = 4.39EE317 pKa = 3.84LQEE320 pKa = 4.11LSRR323 pKa = 11.84RR324 pKa = 11.84AAWSEE329 pKa = 3.74EE330 pKa = 3.74MDD332 pKa = 4.54CYY334 pKa = 10.86PVGLPEE340 pKa = 4.1AFKK343 pKa = 11.24VRR345 pKa = 11.84VITKK349 pKa = 10.1GAVHH353 pKa = 7.27HH354 pKa = 6.45YY355 pKa = 9.81NLARR359 pKa = 11.84RR360 pKa = 11.84WQPAMWEE367 pKa = 4.08PLANHH372 pKa = 6.36PTFEE376 pKa = 4.87LVGTPNCQRR385 pKa = 11.84IMNRR389 pKa = 11.84FLAKK393 pKa = 10.09CDD395 pKa = 4.01PDD397 pKa = 3.93DD398 pKa = 5.29DD399 pKa = 4.47RR400 pKa = 11.84AFTSGDD406 pKa = 3.42YY407 pKa = 11.15ASATDD412 pKa = 4.39YY413 pKa = 11.46LDD415 pKa = 4.3SDD417 pKa = 4.42LSRR420 pKa = 11.84SCLEE424 pKa = 4.05YY425 pKa = 10.13VCNALRR431 pKa = 11.84VPFEE435 pKa = 4.14DD436 pKa = 3.52TVILAEE442 pKa = 4.33ALTDD446 pKa = 3.63HH447 pKa = 7.06RR448 pKa = 11.84LHH450 pKa = 6.47YY451 pKa = 10.4VDD453 pKa = 4.94SEE455 pKa = 4.21GVEE458 pKa = 3.9QVKK461 pKa = 7.79QQRR464 pKa = 11.84RR465 pKa = 11.84GQLMGSPISFPILCLFNAALTRR487 pKa = 11.84YY488 pKa = 8.88ALEE491 pKa = 4.16IASCDD496 pKa = 3.65SVALDD501 pKa = 5.36LEE503 pKa = 4.59DD504 pKa = 6.02LPMLINGDD512 pKa = 4.11DD513 pKa = 4.02LLCRR517 pKa = 11.84TNPLEE522 pKa = 3.87YY523 pKa = 10.49LVWKK527 pKa = 10.26DD528 pKa = 3.07IVNFGGLTPSIGKK541 pKa = 8.67NFRR544 pKa = 11.84HH545 pKa = 5.35RR546 pKa = 11.84TIGTINSEE554 pKa = 3.5MWTFKK559 pKa = 9.85HH560 pKa = 6.32TIHH563 pKa = 6.83GCPANGTDD571 pKa = 2.94TAFYY575 pKa = 9.61QGKK578 pKa = 8.26RR579 pKa = 11.84QPIIEE584 pKa = 4.39IGLVRR589 pKa = 11.84GSVKK593 pKa = 10.44RR594 pKa = 11.84GTINRR599 pKa = 11.84NVDD602 pKa = 3.49EE603 pKa = 5.25LSPFVDD609 pKa = 3.08SSKK612 pKa = 10.53FGKK615 pKa = 10.41SKK617 pKa = 8.53EE618 pKa = 4.12QCWNRR623 pKa = 11.84FLDD626 pKa = 3.67TCPNRR631 pKa = 11.84LTAYY635 pKa = 10.0DD636 pKa = 3.87FLWRR640 pKa = 11.84SVSSDD645 pKa = 2.73ILEE648 pKa = 4.23SLPRR652 pKa = 11.84GMPMCCPTWLGGAGFPLPPLGHH674 pKa = 7.28ALRR677 pKa = 11.84DD678 pKa = 3.57RR679 pKa = 11.84RR680 pKa = 11.84EE681 pKa = 3.58PSAYY685 pKa = 9.47QRR687 pKa = 11.84LLARR691 pKa = 11.84YY692 pKa = 9.27LYY694 pKa = 8.96DD695 pKa = 3.06TYY697 pKa = 11.46GYY699 pKa = 10.71GLGKK703 pKa = 10.1RR704 pKa = 11.84YY705 pKa = 10.64VSMLEE710 pKa = 4.19TGSLPANLVKK720 pKa = 10.59EE721 pKa = 4.32MNTDD725 pKa = 2.9NGIRR729 pKa = 11.84DD730 pKa = 3.68VLGVPKK736 pKa = 9.91PVPEE740 pKa = 4.51SADD743 pKa = 3.38SVDD746 pKa = 4.21GIKK749 pKa = 11.04GNLHH753 pKa = 5.6SEE755 pKa = 4.27KK756 pKa = 10.24TPLTPLPQTHH766 pKa = 6.83FFMCGLINHH775 pKa = 6.21TEE777 pKa = 4.04EE778 pKa = 4.22EE779 pKa = 4.43STGRR783 pKa = 11.84FSCEE787 pKa = 3.08KK788 pKa = 9.17MYY790 pKa = 11.24SRR792 pKa = 11.84LQHH795 pKa = 6.56KK796 pKa = 9.41ALKK799 pKa = 9.78HH800 pKa = 4.86GRR802 pKa = 11.84GPLDD806 pKa = 3.56CKK808 pKa = 10.67FFLPTPPSLKK818 pKa = 10.17LEE820 pKa = 4.13SYY822 pKa = 10.19VFQEE826 pKa = 4.43VVAA829 pKa = 4.65
Molecular weight: 93.49 kDa
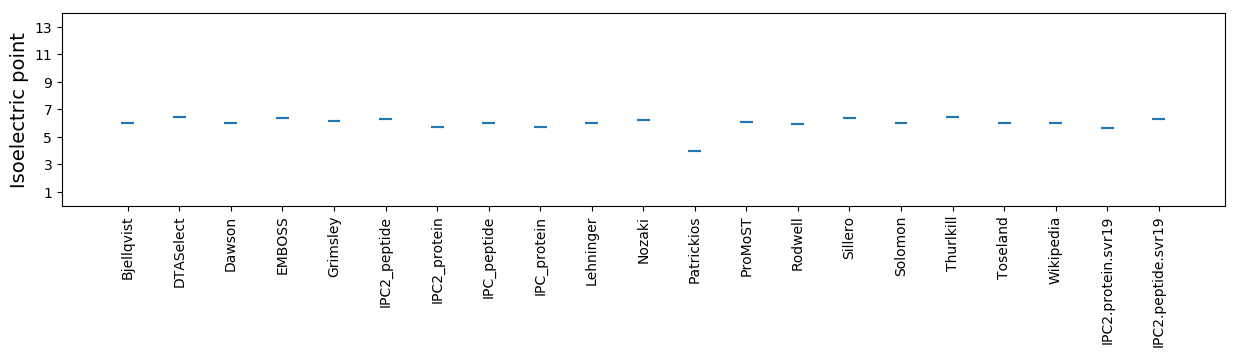
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIM5|A0A1L3KIM5_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 2 OX=1923502 PE=4 SV=1
MM1 pKa = 7.58SPRR4 pKa = 11.84NLFPSYY10 pKa = 11.42GDD12 pKa = 3.34GCSSVNSTEE21 pKa = 3.85NTLASILEE29 pKa = 4.65SPFHH33 pKa = 6.14PAWSAVVEE41 pKa = 4.36GFFGNEE47 pKa = 3.95VVTDD51 pKa = 3.47THH53 pKa = 7.85VDD55 pKa = 3.26EE56 pKa = 4.57LAIYY60 pKa = 10.4EE61 pKa = 4.8DD62 pKa = 4.39GLDD65 pKa = 3.47LSAYY69 pKa = 7.12STSSVEE75 pKa = 4.01QVVRR79 pKa = 11.84CCEE82 pKa = 4.2VYY84 pKa = 10.92SFIYY88 pKa = 9.81DD89 pKa = 3.18QYY91 pKa = 10.67GYY93 pKa = 11.09KK94 pKa = 10.37FLFSPTDD101 pKa = 3.15QRR103 pKa = 11.84LWLEE107 pKa = 4.67LGDD110 pKa = 4.3KK111 pKa = 10.78DD112 pKa = 5.12LLEE115 pKa = 5.36VYY117 pKa = 9.85LKK119 pKa = 10.39WITANILAAVWEE131 pKa = 4.05QDD133 pKa = 3.3EE134 pKa = 4.99LVPLPSSLLDD144 pKa = 3.94FSARR148 pKa = 11.84FPAFGLTTRR157 pKa = 11.84DD158 pKa = 3.23DD159 pKa = 3.68RR160 pKa = 11.84LFRR163 pKa = 11.84SMIRR167 pKa = 11.84SSRR170 pKa = 11.84RR171 pKa = 11.84SKK173 pKa = 9.1KK174 pKa = 6.89QQRR177 pKa = 11.84FVYY180 pKa = 9.7TLYY183 pKa = 10.45QGKK186 pKa = 9.96AGCNAMRR193 pKa = 11.84PEE195 pKa = 4.09LVAAKK200 pKa = 9.9VVSAVEE206 pKa = 4.11TLATPPEE213 pKa = 4.46GEE215 pKa = 4.06EE216 pKa = 4.15SIGILGHH223 pKa = 7.43LITKK227 pKa = 10.06EE228 pKa = 4.06DD229 pKa = 3.26IRR231 pKa = 11.84GQLLRR236 pKa = 11.84TVTEE240 pKa = 4.35IYY242 pKa = 9.18GTGHH246 pKa = 5.11ARR248 pKa = 11.84EE249 pKa = 3.96RR250 pKa = 11.84RR251 pKa = 11.84TPGSRR256 pKa = 11.84RR257 pKa = 11.84MASSKK262 pKa = 11.08SSFQYY267 pKa = 9.93TRR269 pKa = 11.84SKK271 pKa = 11.11GGAHH275 pKa = 6.73EE276 pKa = 4.31FLVDD280 pKa = 3.54NAPASMFIQPRR291 pKa = 11.84ILSGFVVYY299 pKa = 8.69KK300 pKa = 8.71TQVRR304 pKa = 11.84PYY306 pKa = 8.49YY307 pKa = 8.73TTFQAEE313 pKa = 4.31DD314 pKa = 3.39WEE316 pKa = 4.39EE317 pKa = 3.84LQEE320 pKa = 4.11LSRR323 pKa = 11.84RR324 pKa = 11.84AAWSEE329 pKa = 3.74EE330 pKa = 3.74MDD332 pKa = 4.54CYY334 pKa = 10.86PVGLPEE340 pKa = 4.1AFKK343 pKa = 11.24VRR345 pKa = 11.84VITKK349 pKa = 10.1GAVHH353 pKa = 7.27HH354 pKa = 6.45YY355 pKa = 9.81NLARR359 pKa = 11.84RR360 pKa = 11.84WQPAMWEE367 pKa = 4.08PLANHH372 pKa = 6.36PTFEE376 pKa = 4.87LVGTPNCQRR385 pKa = 11.84IMNRR389 pKa = 11.84FLAKK393 pKa = 10.09CDD395 pKa = 4.01PDD397 pKa = 3.93DD398 pKa = 5.29DD399 pKa = 4.47RR400 pKa = 11.84AFTSGDD406 pKa = 3.42YY407 pKa = 11.15ASATDD412 pKa = 4.39YY413 pKa = 11.46LDD415 pKa = 4.3SDD417 pKa = 4.42LSRR420 pKa = 11.84SCLEE424 pKa = 4.05YY425 pKa = 10.13VCNALRR431 pKa = 11.84VPFEE435 pKa = 4.14DD436 pKa = 3.52TVILAEE442 pKa = 4.33ALTDD446 pKa = 3.63HH447 pKa = 7.06RR448 pKa = 11.84LHH450 pKa = 6.47YY451 pKa = 10.4VDD453 pKa = 4.94SEE455 pKa = 4.21GVEE458 pKa = 3.9QVKK461 pKa = 7.79QQRR464 pKa = 11.84RR465 pKa = 11.84GQLMGSPISFPILCLFNAALTRR487 pKa = 11.84YY488 pKa = 8.88ALEE491 pKa = 4.16IASCDD496 pKa = 3.65SVALDD501 pKa = 5.36LEE503 pKa = 4.59DD504 pKa = 6.02LPMLINGDD512 pKa = 4.11DD513 pKa = 4.02LLCRR517 pKa = 11.84TNPLEE522 pKa = 3.87YY523 pKa = 10.49LVWKK527 pKa = 10.26DD528 pKa = 3.07IVNFGGLTPSIGKK541 pKa = 8.67NFRR544 pKa = 11.84HH545 pKa = 5.35RR546 pKa = 11.84TIGTINSEE554 pKa = 3.5MWTFKK559 pKa = 9.85HH560 pKa = 6.32TIHH563 pKa = 6.83GCPANGTDD571 pKa = 2.94TAFYY575 pKa = 9.61QGKK578 pKa = 8.26RR579 pKa = 11.84QPIIEE584 pKa = 4.39IGLVRR589 pKa = 11.84GSVKK593 pKa = 10.44RR594 pKa = 11.84GTINRR599 pKa = 11.84NVDD602 pKa = 3.49EE603 pKa = 5.25LSPFVDD609 pKa = 3.08SSKK612 pKa = 10.53FGKK615 pKa = 10.41SKK617 pKa = 8.53EE618 pKa = 4.12QCWNRR623 pKa = 11.84FLDD626 pKa = 3.67TCPNRR631 pKa = 11.84LTAYY635 pKa = 10.0DD636 pKa = 3.87FLWRR640 pKa = 11.84SVSSDD645 pKa = 2.73ILEE648 pKa = 4.23SLPRR652 pKa = 11.84GMPMCCPTWLGGAGFPLPPLGHH674 pKa = 7.28ALRR677 pKa = 11.84DD678 pKa = 3.57RR679 pKa = 11.84RR680 pKa = 11.84EE681 pKa = 3.58PSAYY685 pKa = 9.47QRR687 pKa = 11.84LLARR691 pKa = 11.84YY692 pKa = 9.27LYY694 pKa = 8.96DD695 pKa = 3.06TYY697 pKa = 11.46GYY699 pKa = 10.71GLGKK703 pKa = 10.1RR704 pKa = 11.84YY705 pKa = 10.64VSMLEE710 pKa = 4.19TGSLPANLVKK720 pKa = 10.59EE721 pKa = 4.32MNTDD725 pKa = 2.9NGIRR729 pKa = 11.84DD730 pKa = 3.68VLGVPKK736 pKa = 9.91PVPEE740 pKa = 4.51SADD743 pKa = 3.38SVDD746 pKa = 4.21GIKK749 pKa = 11.04GNLHH753 pKa = 5.6SEE755 pKa = 4.27KK756 pKa = 10.24TPLTPLPQTHH766 pKa = 6.83FFMCGLINHH775 pKa = 6.21TEE777 pKa = 4.04EE778 pKa = 4.22EE779 pKa = 4.43STGRR783 pKa = 11.84FSCEE787 pKa = 3.08KK788 pKa = 9.17MYY790 pKa = 11.24SRR792 pKa = 11.84LQHH795 pKa = 6.56KK796 pKa = 9.41ALKK799 pKa = 9.78HH800 pKa = 4.86GRR802 pKa = 11.84GPLDD806 pKa = 3.56CKK808 pKa = 10.67FFLPTPPSLKK818 pKa = 10.17LEE820 pKa = 4.13SYY822 pKa = 10.19VFQEE826 pKa = 4.43VVAA829 pKa = 4.65
MM1 pKa = 7.58SPRR4 pKa = 11.84NLFPSYY10 pKa = 11.42GDD12 pKa = 3.34GCSSVNSTEE21 pKa = 3.85NTLASILEE29 pKa = 4.65SPFHH33 pKa = 6.14PAWSAVVEE41 pKa = 4.36GFFGNEE47 pKa = 3.95VVTDD51 pKa = 3.47THH53 pKa = 7.85VDD55 pKa = 3.26EE56 pKa = 4.57LAIYY60 pKa = 10.4EE61 pKa = 4.8DD62 pKa = 4.39GLDD65 pKa = 3.47LSAYY69 pKa = 7.12STSSVEE75 pKa = 4.01QVVRR79 pKa = 11.84CCEE82 pKa = 4.2VYY84 pKa = 10.92SFIYY88 pKa = 9.81DD89 pKa = 3.18QYY91 pKa = 10.67GYY93 pKa = 11.09KK94 pKa = 10.37FLFSPTDD101 pKa = 3.15QRR103 pKa = 11.84LWLEE107 pKa = 4.67LGDD110 pKa = 4.3KK111 pKa = 10.78DD112 pKa = 5.12LLEE115 pKa = 5.36VYY117 pKa = 9.85LKK119 pKa = 10.39WITANILAAVWEE131 pKa = 4.05QDD133 pKa = 3.3EE134 pKa = 4.99LVPLPSSLLDD144 pKa = 3.94FSARR148 pKa = 11.84FPAFGLTTRR157 pKa = 11.84DD158 pKa = 3.23DD159 pKa = 3.68RR160 pKa = 11.84LFRR163 pKa = 11.84SMIRR167 pKa = 11.84SSRR170 pKa = 11.84RR171 pKa = 11.84SKK173 pKa = 9.1KK174 pKa = 6.89QQRR177 pKa = 11.84FVYY180 pKa = 9.7TLYY183 pKa = 10.45QGKK186 pKa = 9.96AGCNAMRR193 pKa = 11.84PEE195 pKa = 4.09LVAAKK200 pKa = 9.9VVSAVEE206 pKa = 4.11TLATPPEE213 pKa = 4.46GEE215 pKa = 4.06EE216 pKa = 4.15SIGILGHH223 pKa = 7.43LITKK227 pKa = 10.06EE228 pKa = 4.06DD229 pKa = 3.26IRR231 pKa = 11.84GQLLRR236 pKa = 11.84TVTEE240 pKa = 4.35IYY242 pKa = 9.18GTGHH246 pKa = 5.11ARR248 pKa = 11.84EE249 pKa = 3.96RR250 pKa = 11.84RR251 pKa = 11.84TPGSRR256 pKa = 11.84RR257 pKa = 11.84MASSKK262 pKa = 11.08SSFQYY267 pKa = 9.93TRR269 pKa = 11.84SKK271 pKa = 11.11GGAHH275 pKa = 6.73EE276 pKa = 4.31FLVDD280 pKa = 3.54NAPASMFIQPRR291 pKa = 11.84ILSGFVVYY299 pKa = 8.69KK300 pKa = 8.71TQVRR304 pKa = 11.84PYY306 pKa = 8.49YY307 pKa = 8.73TTFQAEE313 pKa = 4.31DD314 pKa = 3.39WEE316 pKa = 4.39EE317 pKa = 3.84LQEE320 pKa = 4.11LSRR323 pKa = 11.84RR324 pKa = 11.84AAWSEE329 pKa = 3.74EE330 pKa = 3.74MDD332 pKa = 4.54CYY334 pKa = 10.86PVGLPEE340 pKa = 4.1AFKK343 pKa = 11.24VRR345 pKa = 11.84VITKK349 pKa = 10.1GAVHH353 pKa = 7.27HH354 pKa = 6.45YY355 pKa = 9.81NLARR359 pKa = 11.84RR360 pKa = 11.84WQPAMWEE367 pKa = 4.08PLANHH372 pKa = 6.36PTFEE376 pKa = 4.87LVGTPNCQRR385 pKa = 11.84IMNRR389 pKa = 11.84FLAKK393 pKa = 10.09CDD395 pKa = 4.01PDD397 pKa = 3.93DD398 pKa = 5.29DD399 pKa = 4.47RR400 pKa = 11.84AFTSGDD406 pKa = 3.42YY407 pKa = 11.15ASATDD412 pKa = 4.39YY413 pKa = 11.46LDD415 pKa = 4.3SDD417 pKa = 4.42LSRR420 pKa = 11.84SCLEE424 pKa = 4.05YY425 pKa = 10.13VCNALRR431 pKa = 11.84VPFEE435 pKa = 4.14DD436 pKa = 3.52TVILAEE442 pKa = 4.33ALTDD446 pKa = 3.63HH447 pKa = 7.06RR448 pKa = 11.84LHH450 pKa = 6.47YY451 pKa = 10.4VDD453 pKa = 4.94SEE455 pKa = 4.21GVEE458 pKa = 3.9QVKK461 pKa = 7.79QQRR464 pKa = 11.84RR465 pKa = 11.84GQLMGSPISFPILCLFNAALTRR487 pKa = 11.84YY488 pKa = 8.88ALEE491 pKa = 4.16IASCDD496 pKa = 3.65SVALDD501 pKa = 5.36LEE503 pKa = 4.59DD504 pKa = 6.02LPMLINGDD512 pKa = 4.11DD513 pKa = 4.02LLCRR517 pKa = 11.84TNPLEE522 pKa = 3.87YY523 pKa = 10.49LVWKK527 pKa = 10.26DD528 pKa = 3.07IVNFGGLTPSIGKK541 pKa = 8.67NFRR544 pKa = 11.84HH545 pKa = 5.35RR546 pKa = 11.84TIGTINSEE554 pKa = 3.5MWTFKK559 pKa = 9.85HH560 pKa = 6.32TIHH563 pKa = 6.83GCPANGTDD571 pKa = 2.94TAFYY575 pKa = 9.61QGKK578 pKa = 8.26RR579 pKa = 11.84QPIIEE584 pKa = 4.39IGLVRR589 pKa = 11.84GSVKK593 pKa = 10.44RR594 pKa = 11.84GTINRR599 pKa = 11.84NVDD602 pKa = 3.49EE603 pKa = 5.25LSPFVDD609 pKa = 3.08SSKK612 pKa = 10.53FGKK615 pKa = 10.41SKK617 pKa = 8.53EE618 pKa = 4.12QCWNRR623 pKa = 11.84FLDD626 pKa = 3.67TCPNRR631 pKa = 11.84LTAYY635 pKa = 10.0DD636 pKa = 3.87FLWRR640 pKa = 11.84SVSSDD645 pKa = 2.73ILEE648 pKa = 4.23SLPRR652 pKa = 11.84GMPMCCPTWLGGAGFPLPPLGHH674 pKa = 7.28ALRR677 pKa = 11.84DD678 pKa = 3.57RR679 pKa = 11.84RR680 pKa = 11.84EE681 pKa = 3.58PSAYY685 pKa = 9.47QRR687 pKa = 11.84LLARR691 pKa = 11.84YY692 pKa = 9.27LYY694 pKa = 8.96DD695 pKa = 3.06TYY697 pKa = 11.46GYY699 pKa = 10.71GLGKK703 pKa = 10.1RR704 pKa = 11.84YY705 pKa = 10.64VSMLEE710 pKa = 4.19TGSLPANLVKK720 pKa = 10.59EE721 pKa = 4.32MNTDD725 pKa = 2.9NGIRR729 pKa = 11.84DD730 pKa = 3.68VLGVPKK736 pKa = 9.91PVPEE740 pKa = 4.51SADD743 pKa = 3.38SVDD746 pKa = 4.21GIKK749 pKa = 11.04GNLHH753 pKa = 5.6SEE755 pKa = 4.27KK756 pKa = 10.24TPLTPLPQTHH766 pKa = 6.83FFMCGLINHH775 pKa = 6.21TEE777 pKa = 4.04EE778 pKa = 4.22EE779 pKa = 4.43STGRR783 pKa = 11.84FSCEE787 pKa = 3.08KK788 pKa = 9.17MYY790 pKa = 11.24SRR792 pKa = 11.84LQHH795 pKa = 6.56KK796 pKa = 9.41ALKK799 pKa = 9.78HH800 pKa = 4.86GRR802 pKa = 11.84GPLDD806 pKa = 3.56CKK808 pKa = 10.67FFLPTPPSLKK818 pKa = 10.17LEE820 pKa = 4.13SYY822 pKa = 10.19VFQEE826 pKa = 4.43VVAA829 pKa = 4.65
Molecular weight: 93.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
829 |
829 |
829 |
829.0 |
93.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.273 ± 0.0 | 2.413 ± 0.0 |
5.669 ± 0.0 | 6.514 ± 0.0 |
4.704 ± 0.0 | 6.755 ± 0.0 |
2.292 ± 0.0 | 3.981 ± 0.0 |
3.981 ± 0.0 | 10.374 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.051 ± 0.0 | 3.378 ± 0.0 |
6.031 ± 0.0 | 3.136 ± 0.0 |
6.876 ± 0.0 | 7.841 ± 0.0 |
6.031 ± 0.0 | 6.152 ± 0.0 |
1.568 ± 0.0 | 3.981 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
