
Gokushovirinae Bog8989_22
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; Gokushovirinae; unclassified Gokushovirinae
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
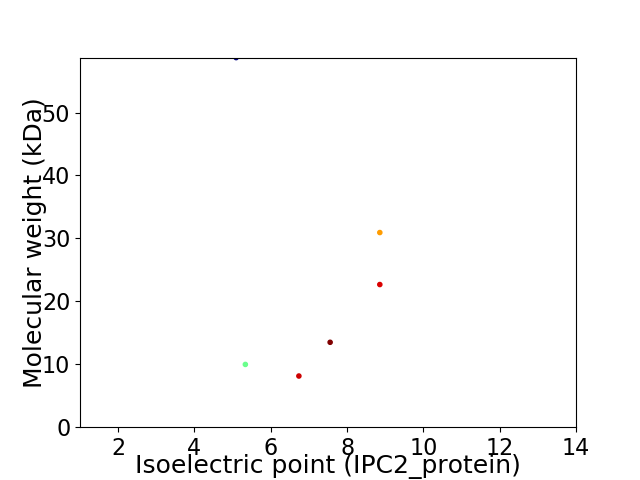
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2UEC9|A0A0G2UEC9_9VIRU DNA binding protein VP5 OS=Gokushovirinae Bog8989_22 OX=1655650 PE=4 SV=1
MM1 pKa = 7.6SSASLISQHH10 pKa = 5.87SFAIAPHH17 pKa = 6.4PKK19 pKa = 9.96LPRR22 pKa = 11.84AIFNRR27 pKa = 11.84TFTHH31 pKa = 6.23KK32 pKa = 6.69TTCNAGMLIPFLIDD46 pKa = 3.77EE47 pKa = 5.35IYY49 pKa = 10.75PGDD52 pKa = 3.85TFKK55 pKa = 11.18LRR57 pKa = 11.84STFLARR63 pKa = 11.84MSTPIYY69 pKa = 10.26PVMDD73 pKa = 3.77NLFIDD78 pKa = 3.79TQFFFIPNRR87 pKa = 11.84LLWTHH92 pKa = 6.54WEE94 pKa = 3.86QFMGYY99 pKa = 8.36QEE101 pKa = 4.63NPGDD105 pKa = 4.4SIDD108 pKa = 3.83YY109 pKa = 7.34TLPTITSPTGGYY121 pKa = 10.02LEE123 pKa = 4.06QTIYY127 pKa = 11.02DD128 pKa = 4.09YY129 pKa = 11.63LGLPTKK135 pKa = 10.33IAGVTHH141 pKa = 5.8QALPLRR147 pKa = 11.84AYY149 pKa = 10.8NKK151 pKa = 9.96VYY153 pKa = 10.69NDD155 pKa = 3.32WYY157 pKa = 10.79RR158 pKa = 11.84DD159 pKa = 3.48QNLQDD164 pKa = 4.32SVVLPNGDD172 pKa = 3.82GPDD175 pKa = 3.47APTDD179 pKa = 3.73YY180 pKa = 11.46VLLPRR185 pKa = 11.84GKK187 pKa = 9.2RR188 pKa = 11.84HH189 pKa = 7.56DD190 pKa = 4.31YY191 pKa = 8.14FTSALPTPQKK201 pKa = 10.58GAAVQLPLGTTAPILSNNTQVNFSDD226 pKa = 3.97EE227 pKa = 4.72AMTSPDD233 pKa = 3.1HH234 pKa = 6.68GLYY237 pKa = 10.15GQEE240 pKa = 3.99GTNLLVLGGSAFGATEE256 pKa = 3.64PLIFGNNTGLYY267 pKa = 9.98ADD269 pKa = 5.02LSSATSATINDD280 pKa = 3.91LRR282 pKa = 11.84LSIATQQYY290 pKa = 9.68LEE292 pKa = 4.12RR293 pKa = 11.84DD294 pKa = 3.49MRR296 pKa = 11.84SGTRR300 pKa = 11.84YY301 pKa = 8.29TEE303 pKa = 3.79VLLGHH308 pKa = 6.99WGIAAQDD315 pKa = 3.37ARR317 pKa = 11.84LQRR320 pKa = 11.84SEE322 pKa = 4.0YY323 pKa = 10.51LGGGSTQINFHH334 pKa = 6.69PVAQTAPSDD343 pKa = 3.48ASGSGEE349 pKa = 3.82EE350 pKa = 4.63SSPQGTLSAFATAVGYY366 pKa = 10.1DD367 pKa = 3.15HH368 pKa = 7.31GFTKK372 pKa = 10.58SFPEE376 pKa = 3.8HH377 pKa = 6.51GFILGLMSIRR387 pKa = 11.84ADD389 pKa = 3.31YY390 pKa = 9.34TYY392 pKa = 10.8QQGAEE397 pKa = 3.91RR398 pKa = 11.84FWFRR402 pKa = 11.84KK403 pKa = 9.7ARR405 pKa = 11.84FDD407 pKa = 4.17LYY409 pKa = 10.78TPEE412 pKa = 3.99FANLGEE418 pKa = 4.11QAVLNQEE425 pKa = 4.66IYY427 pKa = 10.92AQGTSADD434 pKa = 3.91QEE436 pKa = 4.14PFGYY440 pKa = 9.9QEE442 pKa = 3.42AWAEE446 pKa = 3.95LRR448 pKa = 11.84YY449 pKa = 9.99KK450 pKa = 10.38PSRR453 pKa = 11.84ISGYY457 pKa = 9.7FRR459 pKa = 11.84SNATGTLDD467 pKa = 3.09AWHH470 pKa = 7.33LSQEE474 pKa = 4.46FTSPPTLSPAFIIEE488 pKa = 4.47NPPMARR494 pKa = 11.84VEE496 pKa = 4.17AVSDD500 pKa = 3.69VCDD503 pKa = 5.75FIVDD507 pKa = 4.06SNNKK511 pKa = 7.46FHH513 pKa = 7.85CIRR516 pKa = 11.84QIGMYY521 pKa = 9.53SVPGLYY527 pKa = 10.22RR528 pKa = 11.84LL529 pKa = 3.85
MM1 pKa = 7.6SSASLISQHH10 pKa = 5.87SFAIAPHH17 pKa = 6.4PKK19 pKa = 9.96LPRR22 pKa = 11.84AIFNRR27 pKa = 11.84TFTHH31 pKa = 6.23KK32 pKa = 6.69TTCNAGMLIPFLIDD46 pKa = 3.77EE47 pKa = 5.35IYY49 pKa = 10.75PGDD52 pKa = 3.85TFKK55 pKa = 11.18LRR57 pKa = 11.84STFLARR63 pKa = 11.84MSTPIYY69 pKa = 10.26PVMDD73 pKa = 3.77NLFIDD78 pKa = 3.79TQFFFIPNRR87 pKa = 11.84LLWTHH92 pKa = 6.54WEE94 pKa = 3.86QFMGYY99 pKa = 8.36QEE101 pKa = 4.63NPGDD105 pKa = 4.4SIDD108 pKa = 3.83YY109 pKa = 7.34TLPTITSPTGGYY121 pKa = 10.02LEE123 pKa = 4.06QTIYY127 pKa = 11.02DD128 pKa = 4.09YY129 pKa = 11.63LGLPTKK135 pKa = 10.33IAGVTHH141 pKa = 5.8QALPLRR147 pKa = 11.84AYY149 pKa = 10.8NKK151 pKa = 9.96VYY153 pKa = 10.69NDD155 pKa = 3.32WYY157 pKa = 10.79RR158 pKa = 11.84DD159 pKa = 3.48QNLQDD164 pKa = 4.32SVVLPNGDD172 pKa = 3.82GPDD175 pKa = 3.47APTDD179 pKa = 3.73YY180 pKa = 11.46VLLPRR185 pKa = 11.84GKK187 pKa = 9.2RR188 pKa = 11.84HH189 pKa = 7.56DD190 pKa = 4.31YY191 pKa = 8.14FTSALPTPQKK201 pKa = 10.58GAAVQLPLGTTAPILSNNTQVNFSDD226 pKa = 3.97EE227 pKa = 4.72AMTSPDD233 pKa = 3.1HH234 pKa = 6.68GLYY237 pKa = 10.15GQEE240 pKa = 3.99GTNLLVLGGSAFGATEE256 pKa = 3.64PLIFGNNTGLYY267 pKa = 9.98ADD269 pKa = 5.02LSSATSATINDD280 pKa = 3.91LRR282 pKa = 11.84LSIATQQYY290 pKa = 9.68LEE292 pKa = 4.12RR293 pKa = 11.84DD294 pKa = 3.49MRR296 pKa = 11.84SGTRR300 pKa = 11.84YY301 pKa = 8.29TEE303 pKa = 3.79VLLGHH308 pKa = 6.99WGIAAQDD315 pKa = 3.37ARR317 pKa = 11.84LQRR320 pKa = 11.84SEE322 pKa = 4.0YY323 pKa = 10.51LGGGSTQINFHH334 pKa = 6.69PVAQTAPSDD343 pKa = 3.48ASGSGEE349 pKa = 3.82EE350 pKa = 4.63SSPQGTLSAFATAVGYY366 pKa = 10.1DD367 pKa = 3.15HH368 pKa = 7.31GFTKK372 pKa = 10.58SFPEE376 pKa = 3.8HH377 pKa = 6.51GFILGLMSIRR387 pKa = 11.84ADD389 pKa = 3.31YY390 pKa = 9.34TYY392 pKa = 10.8QQGAEE397 pKa = 3.91RR398 pKa = 11.84FWFRR402 pKa = 11.84KK403 pKa = 9.7ARR405 pKa = 11.84FDD407 pKa = 4.17LYY409 pKa = 10.78TPEE412 pKa = 3.99FANLGEE418 pKa = 4.11QAVLNQEE425 pKa = 4.66IYY427 pKa = 10.92AQGTSADD434 pKa = 3.91QEE436 pKa = 4.14PFGYY440 pKa = 9.9QEE442 pKa = 3.42AWAEE446 pKa = 3.95LRR448 pKa = 11.84YY449 pKa = 9.99KK450 pKa = 10.38PSRR453 pKa = 11.84ISGYY457 pKa = 9.7FRR459 pKa = 11.84SNATGTLDD467 pKa = 3.09AWHH470 pKa = 7.33LSQEE474 pKa = 4.46FTSPPTLSPAFIIEE488 pKa = 4.47NPPMARR494 pKa = 11.84VEE496 pKa = 4.17AVSDD500 pKa = 3.69VCDD503 pKa = 5.75FIVDD507 pKa = 4.06SNNKK511 pKa = 7.46FHH513 pKa = 7.85CIRR516 pKa = 11.84QIGMYY521 pKa = 9.53SVPGLYY527 pKa = 10.22RR528 pKa = 11.84LL529 pKa = 3.85
Molecular weight: 58.73 kDa
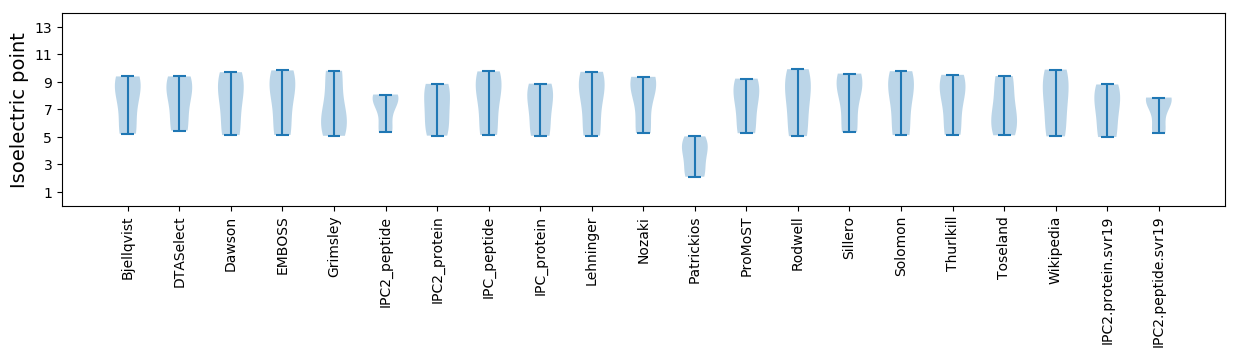
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2UK17|A0A0G2UK17_9VIRU Replication protein VP4 OS=Gokushovirinae Bog8989_22 OX=1655650 PE=4 SV=1
MM1 pKa = 7.53GLFDD5 pKa = 6.2AIGSAVGAGLAYY17 pKa = 10.17EE18 pKa = 4.67GVKK21 pKa = 8.83EE22 pKa = 4.1TNQANKK28 pKa = 10.44DD29 pKa = 3.32INNANNTFSAQQADD43 pKa = 3.43INRR46 pKa = 11.84NFQEE50 pKa = 4.16RR51 pKa = 11.84MSDD54 pKa = 3.11SQYY57 pKa = 10.09QRR59 pKa = 11.84QMADD63 pKa = 2.74MRR65 pKa = 11.84LAGLNPILAASNGGGAGTPSGSAASAGPSPTMQNPMASAVSSAQSVAQTANSIASADD122 pKa = 3.88LTRR125 pKa = 11.84QLALKK130 pKa = 10.45ASAEE134 pKa = 4.27TKK136 pKa = 10.76SIDD139 pKa = 3.55VNSAKK144 pKa = 10.41NAATAPIYY152 pKa = 10.23RR153 pKa = 11.84AAGEE157 pKa = 4.3VTTKK161 pKa = 10.83GLSLWDD167 pKa = 3.62RR168 pKa = 11.84FVPLAWSALSAVPKK182 pKa = 10.43KK183 pKa = 10.9LGGALYY189 pKa = 10.43NGRR192 pKa = 11.84EE193 pKa = 4.5YY194 pKa = 11.27YY195 pKa = 10.16RR196 pKa = 11.84AAHH199 pKa = 5.89NRR201 pKa = 11.84PSAKK205 pKa = 9.66TSWGASGAPSDD216 pKa = 3.87WQPP219 pKa = 2.91
MM1 pKa = 7.53GLFDD5 pKa = 6.2AIGSAVGAGLAYY17 pKa = 10.17EE18 pKa = 4.67GVKK21 pKa = 8.83EE22 pKa = 4.1TNQANKK28 pKa = 10.44DD29 pKa = 3.32INNANNTFSAQQADD43 pKa = 3.43INRR46 pKa = 11.84NFQEE50 pKa = 4.16RR51 pKa = 11.84MSDD54 pKa = 3.11SQYY57 pKa = 10.09QRR59 pKa = 11.84QMADD63 pKa = 2.74MRR65 pKa = 11.84LAGLNPILAASNGGGAGTPSGSAASAGPSPTMQNPMASAVSSAQSVAQTANSIASADD122 pKa = 3.88LTRR125 pKa = 11.84QLALKK130 pKa = 10.45ASAEE134 pKa = 4.27TKK136 pKa = 10.76SIDD139 pKa = 3.55VNSAKK144 pKa = 10.41NAATAPIYY152 pKa = 10.23RR153 pKa = 11.84AAGEE157 pKa = 4.3VTTKK161 pKa = 10.83GLSLWDD167 pKa = 3.62RR168 pKa = 11.84FVPLAWSALSAVPKK182 pKa = 10.43KK183 pKa = 10.9LGGALYY189 pKa = 10.43NGRR192 pKa = 11.84EE193 pKa = 4.5YY194 pKa = 11.27YY195 pKa = 10.16RR196 pKa = 11.84AAHH199 pKa = 5.89NRR201 pKa = 11.84PSAKK205 pKa = 9.66TSWGASGAPSDD216 pKa = 3.87WQPP219 pKa = 2.91
Molecular weight: 22.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1292 |
73 |
529 |
215.3 |
23.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.669 ± 2.15 | 1.316 ± 0.739 |
5.805 ± 0.573 | 3.87 ± 0.268 |
5.108 ± 0.825 | 6.811 ± 0.832 |
2.012 ± 0.446 | 4.954 ± 0.476 |
4.025 ± 0.964 | 9.52 ± 0.844 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.935 ± 0.214 | 4.567 ± 0.675 |
6.347 ± 0.378 | 4.334 ± 0.766 |
5.573 ± 0.939 | 8.359 ± 0.664 |
6.115 ± 0.978 | 4.489 ± 1.007 |
1.006 ± 0.303 | 5.186 ± 0.917 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
