
Salsuginibacillus halophilus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Salsuginibacillus
Average proteome isoelectric point is 5.66
Get precalculated fractions of proteins
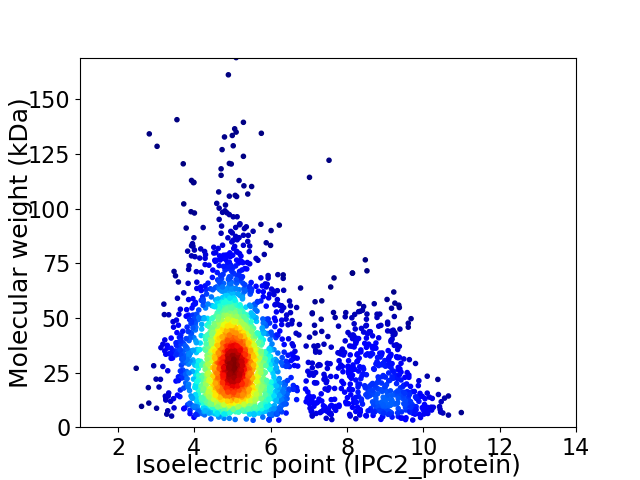
Virtual 2D-PAGE plot for 2921 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P8HQM1|A0A2P8HQM1_9BACI Scaffold Nfu/NifU family protein OS=Salsuginibacillus halophilus OX=517424 GN=B0H94_104108 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 9.97CVKK5 pKa = 10.43GSMVLLFAGVVAGSSQAAAEE25 pKa = 4.15EE26 pKa = 4.57VAFEE30 pKa = 4.22QDD32 pKa = 3.01WKK34 pKa = 10.08TLIDD38 pKa = 4.05DD39 pKa = 4.09RR40 pKa = 11.84NLVEE44 pKa = 5.94DD45 pKa = 4.99ADD47 pKa = 4.19LPGNGEE53 pKa = 4.02IEE55 pKa = 4.23HH56 pKa = 6.49MEE58 pKa = 4.06LFQDD62 pKa = 5.24EE63 pKa = 4.64ILLTDD68 pKa = 4.27GVPFGDD74 pKa = 4.19SFITFVNPEE83 pKa = 4.03NGEE86 pKa = 4.17VIDD89 pKa = 4.63NFTVEE94 pKa = 4.21NPYY97 pKa = 11.29DD98 pKa = 3.85LLIFNQTVDD107 pKa = 3.96DD108 pKa = 4.46YY109 pKa = 11.39IIFDD113 pKa = 4.0GEE115 pKa = 3.72EE116 pKa = 4.18DD117 pKa = 3.59SPYY120 pKa = 10.68DD121 pKa = 3.38YY122 pKa = 11.23TLIYY126 pKa = 9.96SAEE129 pKa = 4.05GDD131 pKa = 3.58KK132 pKa = 10.28VTEE135 pKa = 4.3LKK137 pKa = 10.56DD138 pKa = 3.24AQFIDD143 pKa = 4.8FYY145 pKa = 11.74GEE147 pKa = 3.49NDD149 pKa = 3.39QNILVNGGSGLEE161 pKa = 3.7VRR163 pKa = 11.84DD164 pKa = 4.18FNDD167 pKa = 3.46EE168 pKa = 4.31VIHH171 pKa = 7.21SYY173 pKa = 7.81EE174 pKa = 4.04TEE176 pKa = 3.76ALFEE180 pKa = 4.18FWSTDD185 pKa = 3.14QHH187 pKa = 5.94EE188 pKa = 4.74LNLVLEE194 pKa = 4.76NNMEE198 pKa = 4.1PAIAEE203 pKa = 4.11ADD205 pKa = 3.84NNLKK209 pKa = 9.97PAIIAVDD216 pKa = 3.67NNLEE220 pKa = 4.18QQWEE224 pKa = 4.07ISDD227 pKa = 3.96FPEE230 pKa = 4.78GFADD234 pKa = 3.82EE235 pKa = 5.05PGDD238 pKa = 3.68IRR240 pKa = 11.84SIDD243 pKa = 3.41ASLEE247 pKa = 3.9NTAVIQNRR255 pKa = 11.84EE256 pKa = 4.12NEE258 pKa = 4.27FVTYY262 pKa = 11.15DD263 pKa = 3.18MDD265 pKa = 3.59TGEE268 pKa = 4.3FLTRR272 pKa = 11.84LSDD275 pKa = 3.66YY276 pKa = 11.45SLGNGDD282 pKa = 5.03DD283 pKa = 4.1VVPMIVFDD291 pKa = 4.4DD292 pKa = 5.0RR293 pKa = 11.84IHH295 pKa = 6.69FVDD298 pKa = 4.53EE299 pKa = 4.64SEE301 pKa = 4.18TSNVYY306 pKa = 10.43VYY308 pKa = 10.82DD309 pKa = 3.96EE310 pKa = 3.92NLEE313 pKa = 4.23YY314 pKa = 10.71VDD316 pKa = 3.45HH317 pKa = 7.18HH318 pKa = 5.63EE319 pKa = 4.45TEE321 pKa = 4.1GTEE324 pKa = 3.66IDD326 pKa = 3.68LHH328 pKa = 7.39HH329 pKa = 6.96MYY331 pKa = 9.76MEE333 pKa = 4.25EE334 pKa = 4.39ANYY337 pKa = 10.49QLISDD342 pKa = 4.41YY343 pKa = 11.36EE344 pKa = 4.15NEE346 pKa = 3.99NTKK349 pKa = 10.63LINLSGEE356 pKa = 4.28KK357 pKa = 9.86EE358 pKa = 4.1WTSEE362 pKa = 3.6EE363 pKa = 3.96TLKK366 pKa = 11.1YY367 pKa = 10.85NRR369 pKa = 11.84DD370 pKa = 3.9DD371 pKa = 3.99LSLAGSTLGDD381 pKa = 3.43FAYY384 pKa = 11.26SNFEE388 pKa = 3.87FNNYY392 pKa = 8.39EE393 pKa = 3.86NEE395 pKa = 4.09IYY397 pKa = 11.01LMGVDD402 pKa = 3.61QPMTKK407 pKa = 9.48VHH409 pKa = 6.91EE410 pKa = 4.28IDD412 pKa = 3.0SAYY415 pKa = 10.18FQWTAPMISSDD426 pKa = 3.09GDD428 pKa = 3.51RR429 pKa = 11.84IFFLTNEE436 pKa = 4.17EE437 pKa = 4.8GEE439 pKa = 4.36THH441 pKa = 5.47LTGIEE446 pKa = 4.29RR447 pKa = 11.84STDD450 pKa = 3.26DD451 pKa = 4.9HH452 pKa = 7.42DD453 pKa = 3.84ITPADD458 pKa = 3.93HH459 pKa = 6.89EE460 pKa = 4.56WTIEE464 pKa = 3.9LSQSVDD470 pKa = 3.2DD471 pKa = 4.72HH472 pKa = 7.1SVTEE476 pKa = 3.98SAIYY480 pKa = 10.25IEE482 pKa = 5.18DD483 pKa = 3.58EE484 pKa = 3.93AGEE487 pKa = 4.25RR488 pKa = 11.84IEE490 pKa = 4.61IDD492 pKa = 4.29KK493 pKa = 9.19EE494 pKa = 4.23TADD497 pKa = 4.38QEE499 pKa = 4.87ILIQPADD506 pKa = 3.55GVYY509 pKa = 10.48AAGDD513 pKa = 3.46YY514 pKa = 9.8TLYY517 pKa = 9.83VTDD520 pKa = 4.35EE521 pKa = 4.15LRR523 pKa = 11.84SEE525 pKa = 3.84NGTRR529 pKa = 11.84LNEE532 pKa = 4.18GAAHH536 pKa = 7.04EE537 pKa = 4.53FTVEE541 pKa = 3.61
MM1 pKa = 7.33KK2 pKa = 9.97CVKK5 pKa = 10.43GSMVLLFAGVVAGSSQAAAEE25 pKa = 4.15EE26 pKa = 4.57VAFEE30 pKa = 4.22QDD32 pKa = 3.01WKK34 pKa = 10.08TLIDD38 pKa = 4.05DD39 pKa = 4.09RR40 pKa = 11.84NLVEE44 pKa = 5.94DD45 pKa = 4.99ADD47 pKa = 4.19LPGNGEE53 pKa = 4.02IEE55 pKa = 4.23HH56 pKa = 6.49MEE58 pKa = 4.06LFQDD62 pKa = 5.24EE63 pKa = 4.64ILLTDD68 pKa = 4.27GVPFGDD74 pKa = 4.19SFITFVNPEE83 pKa = 4.03NGEE86 pKa = 4.17VIDD89 pKa = 4.63NFTVEE94 pKa = 4.21NPYY97 pKa = 11.29DD98 pKa = 3.85LLIFNQTVDD107 pKa = 3.96DD108 pKa = 4.46YY109 pKa = 11.39IIFDD113 pKa = 4.0GEE115 pKa = 3.72EE116 pKa = 4.18DD117 pKa = 3.59SPYY120 pKa = 10.68DD121 pKa = 3.38YY122 pKa = 11.23TLIYY126 pKa = 9.96SAEE129 pKa = 4.05GDD131 pKa = 3.58KK132 pKa = 10.28VTEE135 pKa = 4.3LKK137 pKa = 10.56DD138 pKa = 3.24AQFIDD143 pKa = 4.8FYY145 pKa = 11.74GEE147 pKa = 3.49NDD149 pKa = 3.39QNILVNGGSGLEE161 pKa = 3.7VRR163 pKa = 11.84DD164 pKa = 4.18FNDD167 pKa = 3.46EE168 pKa = 4.31VIHH171 pKa = 7.21SYY173 pKa = 7.81EE174 pKa = 4.04TEE176 pKa = 3.76ALFEE180 pKa = 4.18FWSTDD185 pKa = 3.14QHH187 pKa = 5.94EE188 pKa = 4.74LNLVLEE194 pKa = 4.76NNMEE198 pKa = 4.1PAIAEE203 pKa = 4.11ADD205 pKa = 3.84NNLKK209 pKa = 9.97PAIIAVDD216 pKa = 3.67NNLEE220 pKa = 4.18QQWEE224 pKa = 4.07ISDD227 pKa = 3.96FPEE230 pKa = 4.78GFADD234 pKa = 3.82EE235 pKa = 5.05PGDD238 pKa = 3.68IRR240 pKa = 11.84SIDD243 pKa = 3.41ASLEE247 pKa = 3.9NTAVIQNRR255 pKa = 11.84EE256 pKa = 4.12NEE258 pKa = 4.27FVTYY262 pKa = 11.15DD263 pKa = 3.18MDD265 pKa = 3.59TGEE268 pKa = 4.3FLTRR272 pKa = 11.84LSDD275 pKa = 3.66YY276 pKa = 11.45SLGNGDD282 pKa = 5.03DD283 pKa = 4.1VVPMIVFDD291 pKa = 4.4DD292 pKa = 5.0RR293 pKa = 11.84IHH295 pKa = 6.69FVDD298 pKa = 4.53EE299 pKa = 4.64SEE301 pKa = 4.18TSNVYY306 pKa = 10.43VYY308 pKa = 10.82DD309 pKa = 3.96EE310 pKa = 3.92NLEE313 pKa = 4.23YY314 pKa = 10.71VDD316 pKa = 3.45HH317 pKa = 7.18HH318 pKa = 5.63EE319 pKa = 4.45TEE321 pKa = 4.1GTEE324 pKa = 3.66IDD326 pKa = 3.68LHH328 pKa = 7.39HH329 pKa = 6.96MYY331 pKa = 9.76MEE333 pKa = 4.25EE334 pKa = 4.39ANYY337 pKa = 10.49QLISDD342 pKa = 4.41YY343 pKa = 11.36EE344 pKa = 4.15NEE346 pKa = 3.99NTKK349 pKa = 10.63LINLSGEE356 pKa = 4.28KK357 pKa = 9.86EE358 pKa = 4.1WTSEE362 pKa = 3.6EE363 pKa = 3.96TLKK366 pKa = 11.1YY367 pKa = 10.85NRR369 pKa = 11.84DD370 pKa = 3.9DD371 pKa = 3.99LSLAGSTLGDD381 pKa = 3.43FAYY384 pKa = 11.26SNFEE388 pKa = 3.87FNNYY392 pKa = 8.39EE393 pKa = 3.86NEE395 pKa = 4.09IYY397 pKa = 11.01LMGVDD402 pKa = 3.61QPMTKK407 pKa = 9.48VHH409 pKa = 6.91EE410 pKa = 4.28IDD412 pKa = 3.0SAYY415 pKa = 10.18FQWTAPMISSDD426 pKa = 3.09GDD428 pKa = 3.51RR429 pKa = 11.84IFFLTNEE436 pKa = 4.17EE437 pKa = 4.8GEE439 pKa = 4.36THH441 pKa = 5.47LTGIEE446 pKa = 4.29RR447 pKa = 11.84STDD450 pKa = 3.26DD451 pKa = 4.9HH452 pKa = 7.42DD453 pKa = 3.84ITPADD458 pKa = 3.93HH459 pKa = 6.89EE460 pKa = 4.56WTIEE464 pKa = 3.9LSQSVDD470 pKa = 3.2DD471 pKa = 4.72HH472 pKa = 7.1SVTEE476 pKa = 3.98SAIYY480 pKa = 10.25IEE482 pKa = 5.18DD483 pKa = 3.58EE484 pKa = 3.93AGEE487 pKa = 4.25RR488 pKa = 11.84IEE490 pKa = 4.61IDD492 pKa = 4.29KK493 pKa = 9.19EE494 pKa = 4.23TADD497 pKa = 4.38QEE499 pKa = 4.87ILIQPADD506 pKa = 3.55GVYY509 pKa = 10.48AAGDD513 pKa = 3.46YY514 pKa = 9.8TLYY517 pKa = 9.83VTDD520 pKa = 4.35EE521 pKa = 4.15LRR523 pKa = 11.84SEE525 pKa = 3.84NGTRR529 pKa = 11.84LNEE532 pKa = 4.18GAAHH536 pKa = 7.04EE537 pKa = 4.53FTVEE541 pKa = 3.61
Molecular weight: 61.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P8H812|A0A2P8H812_9BACI Ornithine aminotransferase OS=Salsuginibacillus halophilus OX=517424 GN=rocD PE=3 SV=1
MM1 pKa = 8.28RR2 pKa = 11.84MMLFWLLVIAGLSAGWKK19 pKa = 6.69MRR21 pKa = 11.84KK22 pKa = 9.22RR23 pKa = 11.84LVPMLLASKK32 pKa = 10.33RR33 pKa = 11.84FRR35 pKa = 11.84KK36 pKa = 10.14YY37 pKa = 10.51GLQLAFSVPGFRR49 pKa = 11.84RR50 pKa = 11.84LAWRR54 pKa = 11.84MII56 pKa = 3.71
MM1 pKa = 8.28RR2 pKa = 11.84MMLFWLLVIAGLSAGWKK19 pKa = 6.69MRR21 pKa = 11.84KK22 pKa = 9.22RR23 pKa = 11.84LVPMLLASKK32 pKa = 10.33RR33 pKa = 11.84FRR35 pKa = 11.84KK36 pKa = 10.14YY37 pKa = 10.51GLQLAFSVPGFRR49 pKa = 11.84RR50 pKa = 11.84LAWRR54 pKa = 11.84MII56 pKa = 3.71
Molecular weight: 6.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
855502 |
29 |
1530 |
292.9 |
32.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.096 ± 0.062 | 0.622 ± 0.014 |
5.693 ± 0.059 | 8.783 ± 0.055 |
4.137 ± 0.035 | 7.236 ± 0.042 |
2.407 ± 0.027 | 5.867 ± 0.035 |
4.665 ± 0.049 | 9.435 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.911 ± 0.025 | 3.522 ± 0.026 |
3.95 ± 0.028 | 4.107 ± 0.038 |
4.779 ± 0.039 | 5.474 ± 0.031 |
5.651 ± 0.032 | 7.461 ± 0.036 |
1.057 ± 0.015 | 3.148 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
