
Marine Group I thaumarchaeote SCGC AAA799-D07
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Thaumarchaeota incertae sedis; Marine Group I
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
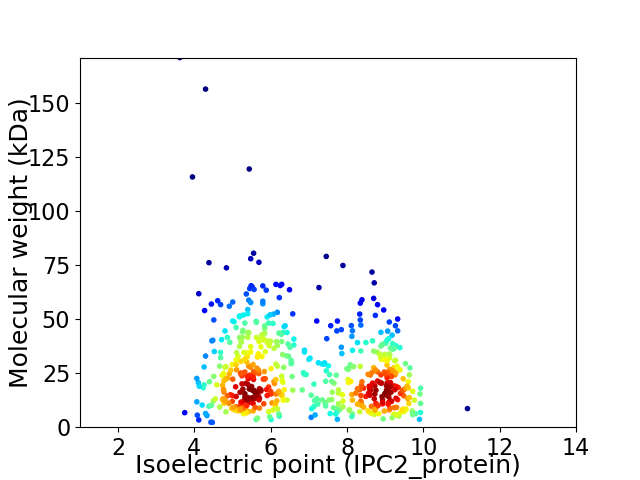
Virtual 2D-PAGE plot for 562 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A087S4J2|A0A087S4J2_9ARCH ATP phosphoribosyltransferase OS=Marine Group I thaumarchaeote SCGC AAA799-D07 OX=1502290 GN=hisG PE=4 SV=1
MM1 pKa = 7.23NNEE4 pKa = 4.48LGRR7 pKa = 11.84KK8 pKa = 7.25ITSLTLMTIMFAGGITVAGSSFAFAPYY35 pKa = 9.34MPEE38 pKa = 3.91AMADD42 pKa = 3.56HH43 pKa = 6.89SNDD46 pKa = 3.39AASSGTLSVSSTHH59 pKa = 5.84VMGGAVLGITVNDD72 pKa = 4.13PSIGATDD79 pKa = 3.58QQIAPPSVEE88 pKa = 3.82YY89 pKa = 10.7DD90 pKa = 3.55GQSVDD95 pKa = 3.58MTQMTDD101 pKa = 3.67GTWQAYY107 pKa = 8.97IVDD110 pKa = 4.07HH111 pKa = 5.74EE112 pKa = 4.91TVVGLDD118 pKa = 2.95GDD120 pKa = 4.15MEE122 pKa = 4.69NDD124 pKa = 3.26NDD126 pKa = 4.35SGFQYY131 pKa = 10.85GVYY134 pKa = 9.96CDD136 pKa = 4.35SGLGAQGVSAGTGNYY151 pKa = 10.09DD152 pKa = 3.29SAGSLDD158 pKa = 4.16SDD160 pKa = 3.36ITTNPSFYY168 pKa = 10.4EE169 pKa = 4.54GWHH172 pKa = 5.94GGGAKK177 pKa = 10.27GKK179 pKa = 10.59GGGFGQTQYY188 pKa = 11.41CQDD191 pKa = 3.55TVDD194 pKa = 4.0AGDD197 pKa = 4.58GIGGGQVKK205 pKa = 9.79FSVLLDD211 pKa = 3.76APTLNTNSGTGTNNSGNRR229 pKa = 11.84NIMVNATTNLSAWPFIQAINFSSTADD255 pKa = 3.07VDD257 pKa = 4.02YY258 pKa = 10.71GAEE261 pKa = 4.15SVTVEE266 pKa = 4.09WGNDD270 pKa = 2.91HH271 pKa = 6.87DD272 pKa = 4.38TSISVDD278 pKa = 3.3RR279 pKa = 11.84DD280 pKa = 3.36IVPDD284 pKa = 3.45NAEE287 pKa = 3.56IHH289 pKa = 5.7LTIVDD294 pKa = 3.91PGLNYY299 pKa = 10.85DD300 pKa = 4.02PTNEE304 pKa = 4.63DD305 pKa = 2.86VWIMDD310 pKa = 4.12AAQEE314 pKa = 3.97TLYY317 pKa = 10.61FYY319 pKa = 11.42NNGSIGEE326 pKa = 4.34KK327 pKa = 9.17NTAQLPTQGASSALTGGEE345 pKa = 3.63ISIDD349 pKa = 3.4GTNNSVIDD357 pKa = 3.72ATTLGNLGCGDD368 pKa = 3.86NCLLSVGGSPKK379 pKa = 10.57NVISTNSSKK388 pKa = 11.07SHH390 pKa = 6.47FGFANITLTEE400 pKa = 4.24TGDD403 pKa = 3.41NTGVFTTFDD412 pKa = 3.5GQTGVSSLKK421 pKa = 10.23SATDD425 pKa = 3.42AGVDD429 pKa = 3.53SQMTFSYY436 pKa = 10.88NGDD439 pKa = 3.46SVTLVIGYY447 pKa = 9.6NDD449 pKa = 3.43ASATFTAGDD458 pKa = 2.93AWLPVEE464 pKa = 4.37DD465 pKa = 4.32AVYY468 pKa = 9.78TITDD472 pKa = 3.38PDD474 pKa = 3.75MNKK477 pKa = 10.09NSLDD481 pKa = 4.82DD482 pKa = 3.72EE483 pKa = 4.54TLDD486 pKa = 4.63IGDD489 pKa = 3.7PHH491 pKa = 8.15DD492 pKa = 5.62RR493 pKa = 11.84IPTIITGSPIHH504 pKa = 6.59VSDD507 pKa = 4.3RR508 pKa = 11.84VSTTARR514 pKa = 11.84EE515 pKa = 4.08HH516 pKa = 7.17SVGQQSLAHH525 pKa = 6.15SSVALSTGSSAANDD539 pKa = 3.33VFYY542 pKa = 8.51TTFASNTTDD551 pKa = 2.64NSKK554 pKa = 10.47RR555 pKa = 11.84IKK557 pKa = 10.69VVVNGIGGGAPTDD570 pKa = 3.76AEE572 pKa = 4.77LNTITWLNVTTDD584 pKa = 3.54IAPSEE589 pKa = 4.04LVDD592 pKa = 4.88LEE594 pKa = 4.48GTVALQYY601 pKa = 10.37DD602 pKa = 3.67ISGIAGALGSTDD614 pKa = 2.34IDD616 pKa = 3.67AYY618 pKa = 11.26VMHH621 pKa = 6.6EE622 pKa = 4.51TVGSTGSEE630 pKa = 3.85PGKK633 pKa = 9.48TMNGTAAIATNVISIVSSGNSASGIFDD660 pKa = 5.14LDD662 pKa = 3.65DD663 pKa = 3.7TTGDD667 pKa = 3.48TAGDD671 pKa = 3.71ATIDD675 pKa = 3.39SAATQKK681 pKa = 10.98VGVNGVEE688 pKa = 4.4DD689 pKa = 3.35AWGVANTNNAVAIVFQITHH708 pKa = 6.74PAGNTLAANGTCAPGGAGHH727 pKa = 7.6DD728 pKa = 3.42KK729 pKa = 11.4GCFGEE734 pKa = 4.28YY735 pKa = 10.14AIALDD740 pKa = 4.27FVNWDD745 pKa = 3.35QDD747 pKa = 3.6NSSKK751 pKa = 10.47SQNGIYY757 pKa = 10.03RR758 pKa = 11.84LEE760 pKa = 4.81AGEE763 pKa = 4.43TGDD766 pKa = 3.4NTGVFTGTVAYY777 pKa = 10.16AILNNSTMITASGDD791 pKa = 3.46PGGSSGFTRR800 pKa = 11.84GGHH803 pKa = 5.78SGNAEE808 pKa = 4.22PEE810 pKa = 4.23PGSAGGVPAPSPSFNGDD827 pKa = 3.16EE828 pKa = 4.4LLVLLGKK835 pKa = 10.33GQTGIDD841 pKa = 3.51APRR844 pKa = 11.84VNINDD849 pKa = 3.54SDD851 pKa = 4.43TVGGTGTTLGVQLDD865 pKa = 4.4ANDD868 pKa = 3.72HH869 pKa = 5.73TGVAEE874 pKa = 4.53FDD876 pKa = 3.73LSSYY880 pKa = 10.64GADD883 pKa = 3.29DD884 pKa = 3.61TATITITDD892 pKa = 4.21LDD894 pKa = 4.5LNQDD898 pKa = 3.33NEE900 pKa = 4.02QRR902 pKa = 11.84EE903 pKa = 4.74VYY905 pKa = 9.09TNSSGTFQIMIDD917 pKa = 4.29DD918 pKa = 5.54DD919 pKa = 5.1DD920 pKa = 4.5SQLLGDD926 pKa = 3.66QVIIEE931 pKa = 4.3TSSNSGVFSGTFVVPNKK948 pKa = 9.97LGKK951 pKa = 10.25DD952 pKa = 3.54MEE954 pKa = 4.35LQYY957 pKa = 11.1YY958 pKa = 10.7DD959 pKa = 4.94SVDD962 pKa = 3.38QSGEE966 pKa = 3.79DD967 pKa = 3.24STTYY971 pKa = 10.77DD972 pKa = 3.3VASIASNLGTVALDD986 pKa = 3.29RR987 pKa = 11.84TSYY990 pKa = 9.56PVPYY994 pKa = 10.32LADD997 pKa = 3.48QLDD1000 pKa = 3.95EE1001 pKa = 4.63GDD1003 pKa = 3.76NTTMTVAKK1011 pKa = 9.49GNVVGTITVTDD1022 pKa = 4.19PDD1024 pKa = 3.56ATADD1028 pKa = 3.62TFTTSTNANSCEE1040 pKa = 3.86AGKK1043 pKa = 10.99VKK1045 pKa = 10.73VKK1047 pKa = 10.77LSGTFVYY1054 pKa = 9.43TAAGSCATAATSTAAQEE1071 pKa = 4.29LGALTEE1077 pKa = 4.31TAQGSGSYY1085 pKa = 10.26EE1086 pKa = 3.63IDD1088 pKa = 3.42FTIGGAACGGSNSGTCQTAQTGVNAVPVNSTSILQVEE1125 pKa = 4.95YY1126 pKa = 10.58IDD1128 pKa = 4.1TADD1131 pKa = 3.53SAGATTTVYY1140 pKa = 11.0DD1141 pKa = 3.52SAIFEE1146 pKa = 4.55LHH1148 pKa = 5.9TATLSVDD1155 pKa = 2.85KK1156 pKa = 10.68DD1157 pKa = 3.99VYY1159 pKa = 11.03ILGQDD1164 pKa = 3.48AVITLSDD1171 pKa = 4.09LDD1173 pKa = 4.63LNLDD1177 pKa = 3.53ADD1179 pKa = 4.58EE1180 pKa = 4.77IEE1182 pKa = 4.48QYY1184 pKa = 10.73DD1185 pKa = 3.9LRR1187 pKa = 11.84LIEE1190 pKa = 4.41WDD1192 pKa = 3.32SDD1194 pKa = 3.87NNSSVLLHH1202 pKa = 6.71SSHH1205 pKa = 6.28CTSGCTLDD1213 pKa = 4.05PSNLTEE1219 pKa = 4.07TGKK1222 pKa = 8.7NTGVFQTVFTIPTSMSDD1239 pKa = 2.95TGAAATSSEE1248 pKa = 4.1MGDD1251 pKa = 3.63EE1252 pKa = 4.5VILTYY1257 pKa = 10.15RR1258 pKa = 11.84DD1259 pKa = 3.32RR1260 pKa = 11.84GLSGEE1265 pKa = 4.11KK1266 pKa = 10.25SYY1268 pKa = 11.87GDD1270 pKa = 4.15DD1271 pKa = 3.77EE1272 pKa = 6.47LDD1274 pKa = 3.57VEE1276 pKa = 4.71ATFSISKK1283 pKa = 10.15FGAIVEE1289 pKa = 4.18LDD1291 pKa = 3.23KK1292 pKa = 11.37AVYY1295 pKa = 9.99DD1296 pKa = 3.77WTDD1299 pKa = 2.9TVYY1302 pKa = 9.41ITVTAPDD1309 pKa = 3.55WNQNSNSEE1317 pKa = 4.21EE1318 pKa = 4.34TIGTSALPINANTRR1332 pKa = 11.84NGKK1335 pKa = 8.98LCSGTNTYY1343 pKa = 8.44TLNEE1347 pKa = 4.05TDD1349 pKa = 3.59EE1350 pKa = 4.38NTGVFDD1356 pKa = 5.34GEE1358 pKa = 3.95IALTGFAHH1366 pKa = 6.74TMATSQSYY1374 pKa = 8.36TPVADD1379 pKa = 3.57ACSGNTDD1386 pKa = 3.77GKK1388 pKa = 10.79LQTAGNMDD1396 pKa = 5.51GITVSYY1402 pKa = 10.04EE1403 pKa = 3.62WVDD1406 pKa = 3.29NSIAIASAIIQWNIGEE1422 pKa = 4.19VEE1424 pKa = 4.19IVDD1427 pKa = 4.29SAVSPNGSTVIRR1439 pKa = 11.84VTDD1442 pKa = 3.37VDD1444 pKa = 3.83EE1445 pKa = 6.37DD1446 pKa = 3.74IDD1448 pKa = 4.07STIVDD1453 pKa = 3.85TFKK1456 pKa = 11.23VDD1458 pKa = 3.33VFSDD1462 pKa = 3.51SDD1464 pKa = 3.64SGGFQATVSEE1474 pKa = 4.48TGEE1477 pKa = 4.08GTGVFEE1483 pKa = 5.21AIIYY1487 pKa = 7.56FTDD1490 pKa = 3.35EE1491 pKa = 4.13TATSGLTLRR1500 pKa = 11.84VAEE1503 pKa = 4.5GDD1505 pKa = 4.01TVTVEE1510 pKa = 4.23YY1511 pKa = 10.77TDD1513 pKa = 3.46VTLPGPDD1520 pKa = 3.54YY1521 pKa = 8.25TTADD1525 pKa = 5.12DD1526 pKa = 3.51ITIAATTTVGTTTPPLEE1543 pKa = 4.3RR1544 pKa = 11.84APAANARR1551 pKa = 11.84VVDD1554 pKa = 3.99AFGSSVAEE1562 pKa = 3.88VSVDD1566 pKa = 3.47QQVQIAADD1574 pKa = 3.51VANAQSKK1581 pKa = 7.96EE1582 pKa = 3.68QSFAYY1587 pKa = 9.96LVQVQDD1593 pKa = 3.86ANGVTVSLAWITGSLNAGQSMSPALSWTPSASGSYY1628 pKa = 8.8TATVFVWEE1636 pKa = 4.51SVDD1639 pKa = 3.6NPVALSPTVTVGIDD1653 pKa = 3.28VVV1655 pKa = 3.56
MM1 pKa = 7.23NNEE4 pKa = 4.48LGRR7 pKa = 11.84KK8 pKa = 7.25ITSLTLMTIMFAGGITVAGSSFAFAPYY35 pKa = 9.34MPEE38 pKa = 3.91AMADD42 pKa = 3.56HH43 pKa = 6.89SNDD46 pKa = 3.39AASSGTLSVSSTHH59 pKa = 5.84VMGGAVLGITVNDD72 pKa = 4.13PSIGATDD79 pKa = 3.58QQIAPPSVEE88 pKa = 3.82YY89 pKa = 10.7DD90 pKa = 3.55GQSVDD95 pKa = 3.58MTQMTDD101 pKa = 3.67GTWQAYY107 pKa = 8.97IVDD110 pKa = 4.07HH111 pKa = 5.74EE112 pKa = 4.91TVVGLDD118 pKa = 2.95GDD120 pKa = 4.15MEE122 pKa = 4.69NDD124 pKa = 3.26NDD126 pKa = 4.35SGFQYY131 pKa = 10.85GVYY134 pKa = 9.96CDD136 pKa = 4.35SGLGAQGVSAGTGNYY151 pKa = 10.09DD152 pKa = 3.29SAGSLDD158 pKa = 4.16SDD160 pKa = 3.36ITTNPSFYY168 pKa = 10.4EE169 pKa = 4.54GWHH172 pKa = 5.94GGGAKK177 pKa = 10.27GKK179 pKa = 10.59GGGFGQTQYY188 pKa = 11.41CQDD191 pKa = 3.55TVDD194 pKa = 4.0AGDD197 pKa = 4.58GIGGGQVKK205 pKa = 9.79FSVLLDD211 pKa = 3.76APTLNTNSGTGTNNSGNRR229 pKa = 11.84NIMVNATTNLSAWPFIQAINFSSTADD255 pKa = 3.07VDD257 pKa = 4.02YY258 pKa = 10.71GAEE261 pKa = 4.15SVTVEE266 pKa = 4.09WGNDD270 pKa = 2.91HH271 pKa = 6.87DD272 pKa = 4.38TSISVDD278 pKa = 3.3RR279 pKa = 11.84DD280 pKa = 3.36IVPDD284 pKa = 3.45NAEE287 pKa = 3.56IHH289 pKa = 5.7LTIVDD294 pKa = 3.91PGLNYY299 pKa = 10.85DD300 pKa = 4.02PTNEE304 pKa = 4.63DD305 pKa = 2.86VWIMDD310 pKa = 4.12AAQEE314 pKa = 3.97TLYY317 pKa = 10.61FYY319 pKa = 11.42NNGSIGEE326 pKa = 4.34KK327 pKa = 9.17NTAQLPTQGASSALTGGEE345 pKa = 3.63ISIDD349 pKa = 3.4GTNNSVIDD357 pKa = 3.72ATTLGNLGCGDD368 pKa = 3.86NCLLSVGGSPKK379 pKa = 10.57NVISTNSSKK388 pKa = 11.07SHH390 pKa = 6.47FGFANITLTEE400 pKa = 4.24TGDD403 pKa = 3.41NTGVFTTFDD412 pKa = 3.5GQTGVSSLKK421 pKa = 10.23SATDD425 pKa = 3.42AGVDD429 pKa = 3.53SQMTFSYY436 pKa = 10.88NGDD439 pKa = 3.46SVTLVIGYY447 pKa = 9.6NDD449 pKa = 3.43ASATFTAGDD458 pKa = 2.93AWLPVEE464 pKa = 4.37DD465 pKa = 4.32AVYY468 pKa = 9.78TITDD472 pKa = 3.38PDD474 pKa = 3.75MNKK477 pKa = 10.09NSLDD481 pKa = 4.82DD482 pKa = 3.72EE483 pKa = 4.54TLDD486 pKa = 4.63IGDD489 pKa = 3.7PHH491 pKa = 8.15DD492 pKa = 5.62RR493 pKa = 11.84IPTIITGSPIHH504 pKa = 6.59VSDD507 pKa = 4.3RR508 pKa = 11.84VSTTARR514 pKa = 11.84EE515 pKa = 4.08HH516 pKa = 7.17SVGQQSLAHH525 pKa = 6.15SSVALSTGSSAANDD539 pKa = 3.33VFYY542 pKa = 8.51TTFASNTTDD551 pKa = 2.64NSKK554 pKa = 10.47RR555 pKa = 11.84IKK557 pKa = 10.69VVVNGIGGGAPTDD570 pKa = 3.76AEE572 pKa = 4.77LNTITWLNVTTDD584 pKa = 3.54IAPSEE589 pKa = 4.04LVDD592 pKa = 4.88LEE594 pKa = 4.48GTVALQYY601 pKa = 10.37DD602 pKa = 3.67ISGIAGALGSTDD614 pKa = 2.34IDD616 pKa = 3.67AYY618 pKa = 11.26VMHH621 pKa = 6.6EE622 pKa = 4.51TVGSTGSEE630 pKa = 3.85PGKK633 pKa = 9.48TMNGTAAIATNVISIVSSGNSASGIFDD660 pKa = 5.14LDD662 pKa = 3.65DD663 pKa = 3.7TTGDD667 pKa = 3.48TAGDD671 pKa = 3.71ATIDD675 pKa = 3.39SAATQKK681 pKa = 10.98VGVNGVEE688 pKa = 4.4DD689 pKa = 3.35AWGVANTNNAVAIVFQITHH708 pKa = 6.74PAGNTLAANGTCAPGGAGHH727 pKa = 7.6DD728 pKa = 3.42KK729 pKa = 11.4GCFGEE734 pKa = 4.28YY735 pKa = 10.14AIALDD740 pKa = 4.27FVNWDD745 pKa = 3.35QDD747 pKa = 3.6NSSKK751 pKa = 10.47SQNGIYY757 pKa = 10.03RR758 pKa = 11.84LEE760 pKa = 4.81AGEE763 pKa = 4.43TGDD766 pKa = 3.4NTGVFTGTVAYY777 pKa = 10.16AILNNSTMITASGDD791 pKa = 3.46PGGSSGFTRR800 pKa = 11.84GGHH803 pKa = 5.78SGNAEE808 pKa = 4.22PEE810 pKa = 4.23PGSAGGVPAPSPSFNGDD827 pKa = 3.16EE828 pKa = 4.4LLVLLGKK835 pKa = 10.33GQTGIDD841 pKa = 3.51APRR844 pKa = 11.84VNINDD849 pKa = 3.54SDD851 pKa = 4.43TVGGTGTTLGVQLDD865 pKa = 4.4ANDD868 pKa = 3.72HH869 pKa = 5.73TGVAEE874 pKa = 4.53FDD876 pKa = 3.73LSSYY880 pKa = 10.64GADD883 pKa = 3.29DD884 pKa = 3.61TATITITDD892 pKa = 4.21LDD894 pKa = 4.5LNQDD898 pKa = 3.33NEE900 pKa = 4.02QRR902 pKa = 11.84EE903 pKa = 4.74VYY905 pKa = 9.09TNSSGTFQIMIDD917 pKa = 4.29DD918 pKa = 5.54DD919 pKa = 5.1DD920 pKa = 4.5SQLLGDD926 pKa = 3.66QVIIEE931 pKa = 4.3TSSNSGVFSGTFVVPNKK948 pKa = 9.97LGKK951 pKa = 10.25DD952 pKa = 3.54MEE954 pKa = 4.35LQYY957 pKa = 11.1YY958 pKa = 10.7DD959 pKa = 4.94SVDD962 pKa = 3.38QSGEE966 pKa = 3.79DD967 pKa = 3.24STTYY971 pKa = 10.77DD972 pKa = 3.3VASIASNLGTVALDD986 pKa = 3.29RR987 pKa = 11.84TSYY990 pKa = 9.56PVPYY994 pKa = 10.32LADD997 pKa = 3.48QLDD1000 pKa = 3.95EE1001 pKa = 4.63GDD1003 pKa = 3.76NTTMTVAKK1011 pKa = 9.49GNVVGTITVTDD1022 pKa = 4.19PDD1024 pKa = 3.56ATADD1028 pKa = 3.62TFTTSTNANSCEE1040 pKa = 3.86AGKK1043 pKa = 10.99VKK1045 pKa = 10.73VKK1047 pKa = 10.77LSGTFVYY1054 pKa = 9.43TAAGSCATAATSTAAQEE1071 pKa = 4.29LGALTEE1077 pKa = 4.31TAQGSGSYY1085 pKa = 10.26EE1086 pKa = 3.63IDD1088 pKa = 3.42FTIGGAACGGSNSGTCQTAQTGVNAVPVNSTSILQVEE1125 pKa = 4.95YY1126 pKa = 10.58IDD1128 pKa = 4.1TADD1131 pKa = 3.53SAGATTTVYY1140 pKa = 11.0DD1141 pKa = 3.52SAIFEE1146 pKa = 4.55LHH1148 pKa = 5.9TATLSVDD1155 pKa = 2.85KK1156 pKa = 10.68DD1157 pKa = 3.99VYY1159 pKa = 11.03ILGQDD1164 pKa = 3.48AVITLSDD1171 pKa = 4.09LDD1173 pKa = 4.63LNLDD1177 pKa = 3.53ADD1179 pKa = 4.58EE1180 pKa = 4.77IEE1182 pKa = 4.48QYY1184 pKa = 10.73DD1185 pKa = 3.9LRR1187 pKa = 11.84LIEE1190 pKa = 4.41WDD1192 pKa = 3.32SDD1194 pKa = 3.87NNSSVLLHH1202 pKa = 6.71SSHH1205 pKa = 6.28CTSGCTLDD1213 pKa = 4.05PSNLTEE1219 pKa = 4.07TGKK1222 pKa = 8.7NTGVFQTVFTIPTSMSDD1239 pKa = 2.95TGAAATSSEE1248 pKa = 4.1MGDD1251 pKa = 3.63EE1252 pKa = 4.5VILTYY1257 pKa = 10.15RR1258 pKa = 11.84DD1259 pKa = 3.32RR1260 pKa = 11.84GLSGEE1265 pKa = 4.11KK1266 pKa = 10.25SYY1268 pKa = 11.87GDD1270 pKa = 4.15DD1271 pKa = 3.77EE1272 pKa = 6.47LDD1274 pKa = 3.57VEE1276 pKa = 4.71ATFSISKK1283 pKa = 10.15FGAIVEE1289 pKa = 4.18LDD1291 pKa = 3.23KK1292 pKa = 11.37AVYY1295 pKa = 9.99DD1296 pKa = 3.77WTDD1299 pKa = 2.9TVYY1302 pKa = 9.41ITVTAPDD1309 pKa = 3.55WNQNSNSEE1317 pKa = 4.21EE1318 pKa = 4.34TIGTSALPINANTRR1332 pKa = 11.84NGKK1335 pKa = 8.98LCSGTNTYY1343 pKa = 8.44TLNEE1347 pKa = 4.05TDD1349 pKa = 3.59EE1350 pKa = 4.38NTGVFDD1356 pKa = 5.34GEE1358 pKa = 3.95IALTGFAHH1366 pKa = 6.74TMATSQSYY1374 pKa = 8.36TPVADD1379 pKa = 3.57ACSGNTDD1386 pKa = 3.77GKK1388 pKa = 10.79LQTAGNMDD1396 pKa = 5.51GITVSYY1402 pKa = 10.04EE1403 pKa = 3.62WVDD1406 pKa = 3.29NSIAIASAIIQWNIGEE1422 pKa = 4.19VEE1424 pKa = 4.19IVDD1427 pKa = 4.29SAVSPNGSTVIRR1439 pKa = 11.84VTDD1442 pKa = 3.37VDD1444 pKa = 3.83EE1445 pKa = 6.37DD1446 pKa = 3.74IDD1448 pKa = 4.07STIVDD1453 pKa = 3.85TFKK1456 pKa = 11.23VDD1458 pKa = 3.33VFSDD1462 pKa = 3.51SDD1464 pKa = 3.64SGGFQATVSEE1474 pKa = 4.48TGEE1477 pKa = 4.08GTGVFEE1483 pKa = 5.21AIIYY1487 pKa = 7.56FTDD1490 pKa = 3.35EE1491 pKa = 4.13TATSGLTLRR1500 pKa = 11.84VAEE1503 pKa = 4.5GDD1505 pKa = 4.01TVTVEE1510 pKa = 4.23YY1511 pKa = 10.77TDD1513 pKa = 3.46VTLPGPDD1520 pKa = 3.54YY1521 pKa = 8.25TTADD1525 pKa = 5.12DD1526 pKa = 3.51ITIAATTTVGTTTPPLEE1543 pKa = 4.3RR1544 pKa = 11.84APAANARR1551 pKa = 11.84VVDD1554 pKa = 3.99AFGSSVAEE1562 pKa = 3.88VSVDD1566 pKa = 3.47QQVQIAADD1574 pKa = 3.51VANAQSKK1581 pKa = 7.96EE1582 pKa = 3.68QSFAYY1587 pKa = 9.96LVQVQDD1593 pKa = 3.86ANGVTVSLAWITGSLNAGQSMSPALSWTPSASGSYY1628 pKa = 8.8TATVFVWEE1636 pKa = 4.51SVDD1639 pKa = 3.6NPVALSPTVTVGIDD1653 pKa = 3.28VVV1655 pKa = 3.56
Molecular weight: 171.2 kDa
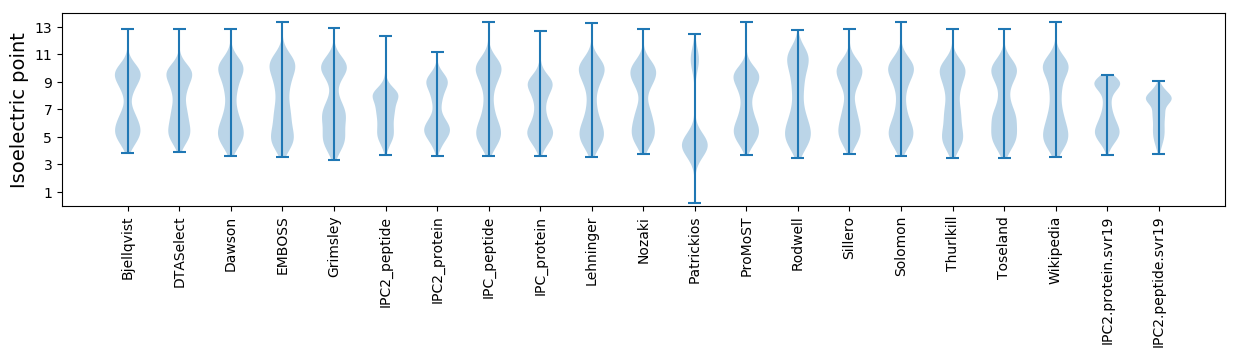
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A087S4D8|A0A087S4D8_9ARCH Uncharacterized protein OS=Marine Group I thaumarchaeote SCGC AAA799-D07 OX=1502290 GN=AAA799D07_00253 PE=4 SV=1
MM1 pKa = 7.59CGKK4 pKa = 10.63LMATSKK10 pKa = 10.69AKK12 pKa = 10.46RR13 pKa = 11.84SAAAKK18 pKa = 9.55KK19 pKa = 9.82AARR22 pKa = 11.84TRR24 pKa = 11.84KK25 pKa = 9.66RR26 pKa = 11.84NAAKK30 pKa = 10.3RR31 pKa = 11.84KK32 pKa = 9.03AAAKK36 pKa = 9.77KK37 pKa = 9.77AAAKK41 pKa = 9.87RR42 pKa = 11.84KK43 pKa = 9.32RR44 pKa = 11.84RR45 pKa = 11.84AAAKK49 pKa = 9.94KK50 pKa = 9.75GGRR53 pKa = 11.84KK54 pKa = 9.42RR55 pKa = 11.84GGAKK59 pKa = 10.09KK60 pKa = 10.42KK61 pKa = 10.39GGKK64 pKa = 8.45KK65 pKa = 9.61RR66 pKa = 11.84KK67 pKa = 8.19AAKK70 pKa = 9.78RR71 pKa = 11.84KK72 pKa = 6.67PAKK75 pKa = 9.55RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 3.56
MM1 pKa = 7.59CGKK4 pKa = 10.63LMATSKK10 pKa = 10.69AKK12 pKa = 10.46RR13 pKa = 11.84SAAAKK18 pKa = 9.55KK19 pKa = 9.82AARR22 pKa = 11.84TRR24 pKa = 11.84KK25 pKa = 9.66RR26 pKa = 11.84NAAKK30 pKa = 10.3RR31 pKa = 11.84KK32 pKa = 9.03AAAKK36 pKa = 9.77KK37 pKa = 9.77AAAKK41 pKa = 9.87RR42 pKa = 11.84KK43 pKa = 9.32RR44 pKa = 11.84RR45 pKa = 11.84AAAKK49 pKa = 9.94KK50 pKa = 9.75GGRR53 pKa = 11.84KK54 pKa = 9.42RR55 pKa = 11.84GGAKK59 pKa = 10.09KK60 pKa = 10.42KK61 pKa = 10.39GGKK64 pKa = 8.45KK65 pKa = 9.61RR66 pKa = 11.84KK67 pKa = 8.19AAKK70 pKa = 9.78RR71 pKa = 11.84KK72 pKa = 6.67PAKK75 pKa = 9.55RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 3.56
Molecular weight: 8.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
133761 |
20 |
1655 |
238.0 |
26.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.02 ± 0.111 | 1.097 ± 0.041 |
5.709 ± 0.09 | 6.503 ± 0.106 |
4.395 ± 0.08 | 6.54 ± 0.105 |
1.844 ± 0.044 | 9.169 ± 0.107 |
8.997 ± 0.159 | 8.745 ± 0.123 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.557 ± 0.048 | 5.367 ± 0.098 |
3.643 ± 0.071 | 2.885 ± 0.058 |
3.52 ± 0.076 | 7.026 ± 0.092 |
5.564 ± 0.104 | 6.393 ± 0.084 |
0.896 ± 0.04 | 3.127 ± 0.066 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
