
Peanut bud necrosis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Tospoviridae; Orthotospovirus; Groundnut bud necrosis orthotospovirus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
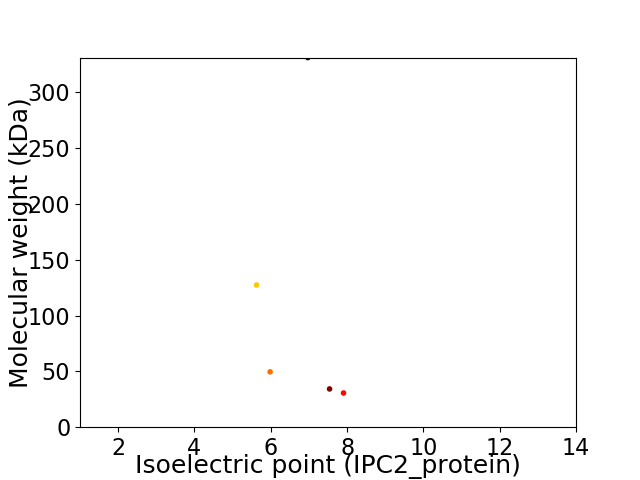
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
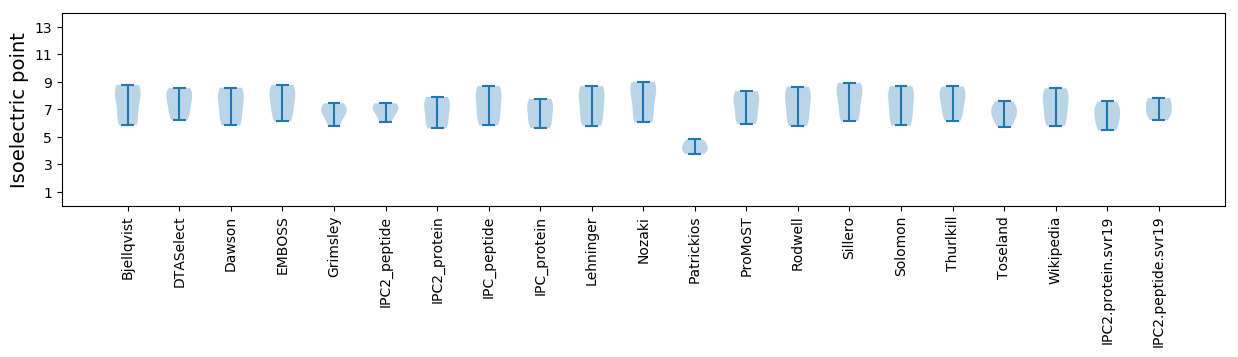
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q84405|Q84405_9VIRU Envelopment polyprotein OS=Peanut bud necrosis virus OX=40687 GN=GPp PE=3 SV=1
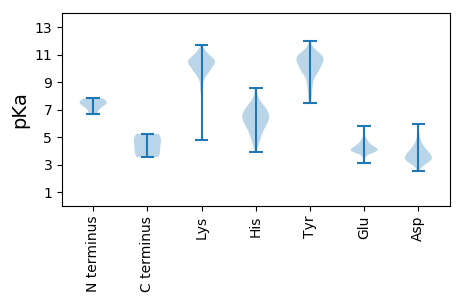
MM1 pKa = 7.46KK2 pKa = 10.1KK3 pKa = 10.47YY4 pKa = 10.74YY5 pKa = 10.82LLVYY9 pKa = 10.52CLGLVSLFFVSEE21 pKa = 4.57VYY23 pKa = 10.88LLNQVDD29 pKa = 3.88NSVQLRR35 pKa = 11.84KK36 pKa = 9.74IQDD39 pKa = 3.33RR40 pKa = 11.84YY41 pKa = 10.97KK42 pKa = 10.9VDD44 pKa = 3.77DD45 pKa = 4.53PEE47 pKa = 5.7DD48 pKa = 3.56LVEE51 pKa = 4.57EE52 pKa = 4.15EE53 pKa = 5.07DD54 pKa = 4.41NIITEE59 pKa = 4.51IIEE62 pKa = 3.96PRR64 pKa = 11.84QKK66 pKa = 10.53KK67 pKa = 10.03LSRR70 pKa = 11.84ILRR73 pKa = 11.84NDD75 pKa = 2.96EE76 pKa = 3.75VTTQSSVSEE85 pKa = 3.91LSCDD89 pKa = 3.08IFEE92 pKa = 5.57KK93 pKa = 9.33RR94 pKa = 11.84HH95 pKa = 5.3CMIKK99 pKa = 10.49GVSDD103 pKa = 4.51FNAHH107 pKa = 4.82YY108 pKa = 10.67QIDD111 pKa = 3.98NGNEE115 pKa = 3.88IISCISNSANIFDD128 pKa = 3.45ICQYY132 pKa = 10.26EE133 pKa = 4.33KK134 pKa = 10.77EE135 pKa = 4.15FKK137 pKa = 10.32KK138 pKa = 10.75IKK140 pKa = 10.05FKK142 pKa = 10.97SFPVVPVLKK151 pKa = 10.85LEE153 pKa = 4.18NKK155 pKa = 9.46KK156 pKa = 10.39VLEE159 pKa = 4.41IGTKK163 pKa = 10.07FFFVDD168 pKa = 4.25KK169 pKa = 10.82SNNPINIDD177 pKa = 3.25PKK179 pKa = 11.31VNLKK183 pKa = 10.52SPTVARR189 pKa = 11.84LSVRR193 pKa = 11.84LSGDD197 pKa = 3.35CKK199 pKa = 10.31INQVSMSSPYY209 pKa = 9.8QIKK212 pKa = 10.28LRR214 pKa = 11.84SEE216 pKa = 4.13EE217 pKa = 4.15NIGILIKK224 pKa = 10.48NVKK227 pKa = 9.43SSKK230 pKa = 10.13SGNIKK235 pKa = 10.45SIAGDD240 pKa = 3.26TTINFKK246 pKa = 10.84PEE248 pKa = 4.02EE249 pKa = 3.96LDD251 pKa = 3.54GNHH254 pKa = 6.69FLLCGDD260 pKa = 4.1KK261 pKa = 10.89SSLIAKK267 pKa = 9.67VDD269 pKa = 3.34IPVRR273 pKa = 11.84NCVSKK278 pKa = 11.44YY279 pKa = 9.76SDD281 pKa = 3.57EE282 pKa = 4.08PKK284 pKa = 10.64KK285 pKa = 10.85IFFCTNFSYY294 pKa = 10.01FKK296 pKa = 9.94WIFVFLMVAFPISWFIWKK314 pKa = 8.58TKK316 pKa = 10.1DD317 pKa = 4.7ALSIWYY323 pKa = 8.99DD324 pKa = 3.33VIGILTYY331 pKa = 10.46PILWTLNWLWPYY343 pKa = 10.52FPLKK347 pKa = 10.55CRR349 pKa = 11.84ICGCFSFLTHH359 pKa = 6.58SCTEE363 pKa = 4.01KK364 pKa = 10.8CVCNQDD370 pKa = 3.89KK371 pKa = 10.74ASKK374 pKa = 10.59DD375 pKa = 3.57HH376 pKa = 6.94TDD378 pKa = 2.88EE379 pKa = 5.59CYY381 pKa = 10.88LFTKK385 pKa = 10.55DD386 pKa = 3.24KK387 pKa = 11.15EE388 pKa = 4.26EE389 pKa = 3.93WRR391 pKa = 11.84KK392 pKa = 8.18LTLIQQFQFIINTKK406 pKa = 10.16LSTNFLVFVTKK417 pKa = 10.41MILASILISYY427 pKa = 9.3IPSSIALKK435 pKa = 10.35QPNICVEE442 pKa = 3.72KK443 pKa = 10.51CYY445 pKa = 11.27YY446 pKa = 10.32NLNLDD451 pKa = 4.1SLTTDD456 pKa = 2.98KK457 pKa = 11.33FGMTDD462 pKa = 3.42NGYY465 pKa = 7.5EE466 pKa = 4.01TCEE469 pKa = 3.93CSIGNVITEE478 pKa = 4.0TVYY481 pKa = 10.83RR482 pKa = 11.84SGVPMSRR489 pKa = 11.84ATSLNDD495 pKa = 3.36CVLGSDD501 pKa = 4.27LCMVSNNQAQNLFACRR517 pKa = 11.84NGCNSLSSIKK527 pKa = 10.42NIPDD531 pKa = 3.27TKK533 pKa = 10.63FNKK536 pKa = 9.87LYY538 pKa = 10.44KK539 pKa = 9.95GQSFKK544 pKa = 11.72GNLTSLKK551 pKa = 9.23IANRR555 pKa = 11.84LRR557 pKa = 11.84EE558 pKa = 4.28GYY560 pKa = 8.76MDD562 pKa = 4.73SPTEE566 pKa = 4.08SKK568 pKa = 10.49ILEE571 pKa = 4.18EE572 pKa = 4.33EE573 pKa = 4.11SAKK576 pKa = 10.38EE577 pKa = 3.71YY578 pKa = 11.07KK579 pKa = 9.96FYY581 pKa = 11.29KK582 pKa = 9.52SLKK585 pKa = 9.71VDD587 pKa = 5.4DD588 pKa = 5.0IPPEE592 pKa = 4.0NLLPRR597 pKa = 11.84QSLVFSTEE605 pKa = 3.11VDD607 pKa = 3.09GKK609 pKa = 10.74YY610 pKa = 9.97RR611 pKa = 11.84YY612 pKa = 9.71LLEE615 pKa = 4.2MDD617 pKa = 3.71IKK619 pKa = 11.17SNTGSVYY626 pKa = 10.92LLNDD630 pKa = 4.29DD631 pKa = 4.2ATHH634 pKa = 6.64SPMEE638 pKa = 3.57FMIYY642 pKa = 9.48IKK644 pKa = 10.62SVGVEE649 pKa = 3.31YY650 pKa = 10.6DD651 pKa = 3.11VRR653 pKa = 11.84YY654 pKa = 9.93KK655 pKa = 11.16YY656 pKa = 9.83STAKK660 pKa = 10.07IDD662 pKa = 3.86TTVSDD667 pKa = 4.23YY668 pKa = 11.57LVTCTGKK675 pKa = 10.63CADD678 pKa = 5.18CIKK681 pKa = 10.37QKK683 pKa = 10.68PKK685 pKa = 10.93VGVLDD690 pKa = 4.06FCVTPTSWWGCEE702 pKa = 3.82EE703 pKa = 4.53LGCLAINEE711 pKa = 4.23GSICGHH717 pKa = 5.92CTNIYY722 pKa = 10.33DD723 pKa = 4.93LSSLVNIYY731 pKa = 10.07QVVEE735 pKa = 3.98SHH737 pKa = 5.44VTAEE741 pKa = 4.25VCVKK745 pKa = 10.75SLDD748 pKa = 3.85GYY750 pKa = 9.1SCKK753 pKa = 10.14KK754 pKa = 10.17HH755 pKa = 6.52SDD757 pKa = 3.26RR758 pKa = 11.84SPIQTDD764 pKa = 4.38YY765 pKa = 10.95YY766 pKa = 10.08QLDD769 pKa = 3.74MSIDD773 pKa = 3.46LHH775 pKa = 7.51NDD777 pKa = 2.92YY778 pKa = 10.78MSTDD782 pKa = 3.11KK783 pKa = 11.27LFAVTKK789 pKa = 7.15QQKK792 pKa = 9.81ILTGNIADD800 pKa = 4.66LGDD803 pKa = 3.62FAGSSFGHH811 pKa = 5.51PQITIDD817 pKa = 5.04GIPLSVPATLSQNDD831 pKa = 4.03FTWSCSAVGDD841 pKa = 3.62KK842 pKa = 10.46KK843 pKa = 11.66VNIKK847 pKa = 10.15QCGLYY852 pKa = 9.02TYY854 pKa = 10.59SAIYY858 pKa = 9.9VLSPSKK864 pKa = 10.66DD865 pKa = 3.43VSHH868 pKa = 7.56LDD870 pKa = 3.45EE871 pKa = 5.03NSNKK875 pKa = 10.45LYY877 pKa = 9.75MEE879 pKa = 4.61KK880 pKa = 10.79DD881 pKa = 3.46FLVGKK886 pKa = 10.29LKK888 pKa = 10.36MVVDD892 pKa = 4.18MPKK895 pKa = 10.84EE896 pKa = 3.97MFKK899 pKa = 10.71KK900 pKa = 10.41IPTKK904 pKa = 10.41PIISEE909 pKa = 3.83AKK911 pKa = 8.95MICSGCSQCAVGIDD925 pKa = 3.78CNITYY930 pKa = 9.74TSDD933 pKa = 3.11TTFSARR939 pKa = 11.84LMMDD943 pKa = 3.59TCSFKK948 pKa = 10.77SDD950 pKa = 3.37QLGTFLGPNEE960 pKa = 4.33KK961 pKa = 10.24SIKK964 pKa = 9.84AYY966 pKa = 10.24CSEE969 pKa = 4.41EE970 pKa = 3.89ITDD973 pKa = 4.37KK974 pKa = 10.92SLKK977 pKa = 10.29LIPEE981 pKa = 4.46DD982 pKa = 4.01QEE984 pKa = 4.11EE985 pKa = 4.64LTVDD989 pKa = 3.28IQVDD993 pKa = 3.75EE994 pKa = 4.65FTQVDD999 pKa = 3.9QDD1001 pKa = 4.34TIIHH1005 pKa = 6.59FDD1007 pKa = 3.87DD1008 pKa = 5.37KK1009 pKa = 11.3SAHH1012 pKa = 6.98DD1013 pKa = 4.17EE1014 pKa = 4.3NKK1016 pKa = 9.65HH1017 pKa = 6.34HH1018 pKa = 7.3SDD1020 pKa = 3.33TSISSLWDD1028 pKa = 3.48WIKK1031 pKa = 11.43APFNWIASFFGSFFDD1046 pKa = 3.64LVRR1049 pKa = 11.84IILVIVAACVGLYY1062 pKa = 9.71IVSSIFKK1069 pKa = 10.52LSRR1072 pKa = 11.84TYY1074 pKa = 11.0YY1075 pKa = 8.94VDD1077 pKa = 3.07KK1078 pKa = 10.72RR1079 pKa = 11.84RR1080 pKa = 11.84QKK1082 pKa = 11.05LEE1084 pKa = 3.82DD1085 pKa = 4.69AIEE1088 pKa = 4.28SVEE1091 pKa = 4.65SSVLLANYY1099 pKa = 8.84TGIDD1103 pKa = 3.25QTRR1106 pKa = 11.84KK1107 pKa = 9.77RR1108 pKa = 11.84KK1109 pKa = 9.82SPPKK1113 pKa = 10.42GYY1115 pKa = 10.48DD1116 pKa = 3.1FSLDD1120 pKa = 3.12II1121 pKa = 5.22
MM1 pKa = 7.46KK2 pKa = 10.1KK3 pKa = 10.47YY4 pKa = 10.74YY5 pKa = 10.82LLVYY9 pKa = 10.52CLGLVSLFFVSEE21 pKa = 4.57VYY23 pKa = 10.88LLNQVDD29 pKa = 3.88NSVQLRR35 pKa = 11.84KK36 pKa = 9.74IQDD39 pKa = 3.33RR40 pKa = 11.84YY41 pKa = 10.97KK42 pKa = 10.9VDD44 pKa = 3.77DD45 pKa = 4.53PEE47 pKa = 5.7DD48 pKa = 3.56LVEE51 pKa = 4.57EE52 pKa = 4.15EE53 pKa = 5.07DD54 pKa = 4.41NIITEE59 pKa = 4.51IIEE62 pKa = 3.96PRR64 pKa = 11.84QKK66 pKa = 10.53KK67 pKa = 10.03LSRR70 pKa = 11.84ILRR73 pKa = 11.84NDD75 pKa = 2.96EE76 pKa = 3.75VTTQSSVSEE85 pKa = 3.91LSCDD89 pKa = 3.08IFEE92 pKa = 5.57KK93 pKa = 9.33RR94 pKa = 11.84HH95 pKa = 5.3CMIKK99 pKa = 10.49GVSDD103 pKa = 4.51FNAHH107 pKa = 4.82YY108 pKa = 10.67QIDD111 pKa = 3.98NGNEE115 pKa = 3.88IISCISNSANIFDD128 pKa = 3.45ICQYY132 pKa = 10.26EE133 pKa = 4.33KK134 pKa = 10.77EE135 pKa = 4.15FKK137 pKa = 10.32KK138 pKa = 10.75IKK140 pKa = 10.05FKK142 pKa = 10.97SFPVVPVLKK151 pKa = 10.85LEE153 pKa = 4.18NKK155 pKa = 9.46KK156 pKa = 10.39VLEE159 pKa = 4.41IGTKK163 pKa = 10.07FFFVDD168 pKa = 4.25KK169 pKa = 10.82SNNPINIDD177 pKa = 3.25PKK179 pKa = 11.31VNLKK183 pKa = 10.52SPTVARR189 pKa = 11.84LSVRR193 pKa = 11.84LSGDD197 pKa = 3.35CKK199 pKa = 10.31INQVSMSSPYY209 pKa = 9.8QIKK212 pKa = 10.28LRR214 pKa = 11.84SEE216 pKa = 4.13EE217 pKa = 4.15NIGILIKK224 pKa = 10.48NVKK227 pKa = 9.43SSKK230 pKa = 10.13SGNIKK235 pKa = 10.45SIAGDD240 pKa = 3.26TTINFKK246 pKa = 10.84PEE248 pKa = 4.02EE249 pKa = 3.96LDD251 pKa = 3.54GNHH254 pKa = 6.69FLLCGDD260 pKa = 4.1KK261 pKa = 10.89SSLIAKK267 pKa = 9.67VDD269 pKa = 3.34IPVRR273 pKa = 11.84NCVSKK278 pKa = 11.44YY279 pKa = 9.76SDD281 pKa = 3.57EE282 pKa = 4.08PKK284 pKa = 10.64KK285 pKa = 10.85IFFCTNFSYY294 pKa = 10.01FKK296 pKa = 9.94WIFVFLMVAFPISWFIWKK314 pKa = 8.58TKK316 pKa = 10.1DD317 pKa = 4.7ALSIWYY323 pKa = 8.99DD324 pKa = 3.33VIGILTYY331 pKa = 10.46PILWTLNWLWPYY343 pKa = 10.52FPLKK347 pKa = 10.55CRR349 pKa = 11.84ICGCFSFLTHH359 pKa = 6.58SCTEE363 pKa = 4.01KK364 pKa = 10.8CVCNQDD370 pKa = 3.89KK371 pKa = 10.74ASKK374 pKa = 10.59DD375 pKa = 3.57HH376 pKa = 6.94TDD378 pKa = 2.88EE379 pKa = 5.59CYY381 pKa = 10.88LFTKK385 pKa = 10.55DD386 pKa = 3.24KK387 pKa = 11.15EE388 pKa = 4.26EE389 pKa = 3.93WRR391 pKa = 11.84KK392 pKa = 8.18LTLIQQFQFIINTKK406 pKa = 10.16LSTNFLVFVTKK417 pKa = 10.41MILASILISYY427 pKa = 9.3IPSSIALKK435 pKa = 10.35QPNICVEE442 pKa = 3.72KK443 pKa = 10.51CYY445 pKa = 11.27YY446 pKa = 10.32NLNLDD451 pKa = 4.1SLTTDD456 pKa = 2.98KK457 pKa = 11.33FGMTDD462 pKa = 3.42NGYY465 pKa = 7.5EE466 pKa = 4.01TCEE469 pKa = 3.93CSIGNVITEE478 pKa = 4.0TVYY481 pKa = 10.83RR482 pKa = 11.84SGVPMSRR489 pKa = 11.84ATSLNDD495 pKa = 3.36CVLGSDD501 pKa = 4.27LCMVSNNQAQNLFACRR517 pKa = 11.84NGCNSLSSIKK527 pKa = 10.42NIPDD531 pKa = 3.27TKK533 pKa = 10.63FNKK536 pKa = 9.87LYY538 pKa = 10.44KK539 pKa = 9.95GQSFKK544 pKa = 11.72GNLTSLKK551 pKa = 9.23IANRR555 pKa = 11.84LRR557 pKa = 11.84EE558 pKa = 4.28GYY560 pKa = 8.76MDD562 pKa = 4.73SPTEE566 pKa = 4.08SKK568 pKa = 10.49ILEE571 pKa = 4.18EE572 pKa = 4.33EE573 pKa = 4.11SAKK576 pKa = 10.38EE577 pKa = 3.71YY578 pKa = 11.07KK579 pKa = 9.96FYY581 pKa = 11.29KK582 pKa = 9.52SLKK585 pKa = 9.71VDD587 pKa = 5.4DD588 pKa = 5.0IPPEE592 pKa = 4.0NLLPRR597 pKa = 11.84QSLVFSTEE605 pKa = 3.11VDD607 pKa = 3.09GKK609 pKa = 10.74YY610 pKa = 9.97RR611 pKa = 11.84YY612 pKa = 9.71LLEE615 pKa = 4.2MDD617 pKa = 3.71IKK619 pKa = 11.17SNTGSVYY626 pKa = 10.92LLNDD630 pKa = 4.29DD631 pKa = 4.2ATHH634 pKa = 6.64SPMEE638 pKa = 3.57FMIYY642 pKa = 9.48IKK644 pKa = 10.62SVGVEE649 pKa = 3.31YY650 pKa = 10.6DD651 pKa = 3.11VRR653 pKa = 11.84YY654 pKa = 9.93KK655 pKa = 11.16YY656 pKa = 9.83STAKK660 pKa = 10.07IDD662 pKa = 3.86TTVSDD667 pKa = 4.23YY668 pKa = 11.57LVTCTGKK675 pKa = 10.63CADD678 pKa = 5.18CIKK681 pKa = 10.37QKK683 pKa = 10.68PKK685 pKa = 10.93VGVLDD690 pKa = 4.06FCVTPTSWWGCEE702 pKa = 3.82EE703 pKa = 4.53LGCLAINEE711 pKa = 4.23GSICGHH717 pKa = 5.92CTNIYY722 pKa = 10.33DD723 pKa = 4.93LSSLVNIYY731 pKa = 10.07QVVEE735 pKa = 3.98SHH737 pKa = 5.44VTAEE741 pKa = 4.25VCVKK745 pKa = 10.75SLDD748 pKa = 3.85GYY750 pKa = 9.1SCKK753 pKa = 10.14KK754 pKa = 10.17HH755 pKa = 6.52SDD757 pKa = 3.26RR758 pKa = 11.84SPIQTDD764 pKa = 4.38YY765 pKa = 10.95YY766 pKa = 10.08QLDD769 pKa = 3.74MSIDD773 pKa = 3.46LHH775 pKa = 7.51NDD777 pKa = 2.92YY778 pKa = 10.78MSTDD782 pKa = 3.11KK783 pKa = 11.27LFAVTKK789 pKa = 7.15QQKK792 pKa = 9.81ILTGNIADD800 pKa = 4.66LGDD803 pKa = 3.62FAGSSFGHH811 pKa = 5.51PQITIDD817 pKa = 5.04GIPLSVPATLSQNDD831 pKa = 4.03FTWSCSAVGDD841 pKa = 3.62KK842 pKa = 10.46KK843 pKa = 11.66VNIKK847 pKa = 10.15QCGLYY852 pKa = 9.02TYY854 pKa = 10.59SAIYY858 pKa = 9.9VLSPSKK864 pKa = 10.66DD865 pKa = 3.43VSHH868 pKa = 7.56LDD870 pKa = 3.45EE871 pKa = 5.03NSNKK875 pKa = 10.45LYY877 pKa = 9.75MEE879 pKa = 4.61KK880 pKa = 10.79DD881 pKa = 3.46FLVGKK886 pKa = 10.29LKK888 pKa = 10.36MVVDD892 pKa = 4.18MPKK895 pKa = 10.84EE896 pKa = 3.97MFKK899 pKa = 10.71KK900 pKa = 10.41IPTKK904 pKa = 10.41PIISEE909 pKa = 3.83AKK911 pKa = 8.95MICSGCSQCAVGIDD925 pKa = 3.78CNITYY930 pKa = 9.74TSDD933 pKa = 3.11TTFSARR939 pKa = 11.84LMMDD943 pKa = 3.59TCSFKK948 pKa = 10.77SDD950 pKa = 3.37QLGTFLGPNEE960 pKa = 4.33KK961 pKa = 10.24SIKK964 pKa = 9.84AYY966 pKa = 10.24CSEE969 pKa = 4.41EE970 pKa = 3.89ITDD973 pKa = 4.37KK974 pKa = 10.92SLKK977 pKa = 10.29LIPEE981 pKa = 4.46DD982 pKa = 4.01QEE984 pKa = 4.11EE985 pKa = 4.64LTVDD989 pKa = 3.28IQVDD993 pKa = 3.75EE994 pKa = 4.65FTQVDD999 pKa = 3.9QDD1001 pKa = 4.34TIIHH1005 pKa = 6.59FDD1007 pKa = 3.87DD1008 pKa = 5.37KK1009 pKa = 11.3SAHH1012 pKa = 6.98DD1013 pKa = 4.17EE1014 pKa = 4.3NKK1016 pKa = 9.65HH1017 pKa = 6.34HH1018 pKa = 7.3SDD1020 pKa = 3.33TSISSLWDD1028 pKa = 3.48WIKK1031 pKa = 11.43APFNWIASFFGSFFDD1046 pKa = 3.64LVRR1049 pKa = 11.84IILVIVAACVGLYY1062 pKa = 9.71IVSSIFKK1069 pKa = 10.52LSRR1072 pKa = 11.84TYY1074 pKa = 11.0YY1075 pKa = 8.94VDD1077 pKa = 3.07KK1078 pKa = 10.72RR1079 pKa = 11.84RR1080 pKa = 11.84QKK1082 pKa = 11.05LEE1084 pKa = 3.82DD1085 pKa = 4.69AIEE1088 pKa = 4.28SVEE1091 pKa = 4.65SSVLLANYY1099 pKa = 8.84TGIDD1103 pKa = 3.25QTRR1106 pKa = 11.84KK1107 pKa = 9.77RR1108 pKa = 11.84KK1109 pKa = 9.82SPPKK1113 pKa = 10.42GYY1115 pKa = 10.48DD1116 pKa = 3.1FSLDD1120 pKa = 3.12II1121 pKa = 5.22
Molecular weight: 127.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q84404|Q84404_9VIRU Non-structural protein OS=Peanut bud necrosis virus OX=40687 GN=NSm PE=4 SV=1
MM1 pKa = 7.52SNVKK5 pKa = 9.98QLTEE9 pKa = 4.08KK10 pKa = 10.51KK11 pKa = 10.05IKK13 pKa = 9.93EE14 pKa = 4.02LLAGGSADD22 pKa = 4.03VEE24 pKa = 4.45IEE26 pKa = 4.46TEE28 pKa = 4.09DD29 pKa = 3.57STPGFSFKK37 pKa = 10.96AFYY40 pKa = 8.24DD41 pKa = 3.67TNKK44 pKa = 10.16NLEE47 pKa = 4.03ITFTNCLNILKK58 pKa = 9.92CRR60 pKa = 11.84KK61 pKa = 9.63QIFAACKK68 pKa = 7.45SGKK71 pKa = 9.77YY72 pKa = 7.99VFCGKK77 pKa = 9.11TIVATNTDD85 pKa = 3.75VGPDD89 pKa = 2.87DD90 pKa = 3.44WTFKK94 pKa = 9.6RR95 pKa = 11.84TEE97 pKa = 3.6AFIRR101 pKa = 11.84TKK103 pKa = 9.04MASMVEE109 pKa = 3.99KK110 pKa = 10.61SKK112 pKa = 11.51NDD114 pKa = 3.46AAKK117 pKa = 10.37QEE119 pKa = 4.19MYY121 pKa = 10.86NKK123 pKa = 9.67IMEE126 pKa = 4.76LPLVAAYY133 pKa = 9.82GLNVPASFDD142 pKa = 3.51TCALRR147 pKa = 11.84MMLCIGGPLPLLSSMTGLAPIIFPLAYY174 pKa = 8.37YY175 pKa = 10.6QNVKK179 pKa = 10.31KK180 pKa = 10.66EE181 pKa = 3.8KK182 pKa = 10.6LGIKK186 pKa = 10.06NFSTYY191 pKa = 9.83EE192 pKa = 3.92QVCKK196 pKa = 9.96VAKK199 pKa = 10.09VLSASQIEE207 pKa = 4.69FKK209 pKa = 11.32NEE211 pKa = 3.41LEE213 pKa = 4.25EE214 pKa = 4.18MFKK217 pKa = 10.96SAVKK221 pKa = 10.36LLSEE225 pKa = 4.79SNPGTASSISLKK237 pKa = 10.73KK238 pKa = 10.13YY239 pKa = 10.29DD240 pKa = 3.9EE241 pKa = 4.12QVKK244 pKa = 10.65YY245 pKa = 9.75MDD247 pKa = 3.99KK248 pKa = 11.02AFSASLSMDD257 pKa = 3.49DD258 pKa = 4.54YY259 pKa = 11.95GEE261 pKa = 4.16HH262 pKa = 6.2SKK264 pKa = 10.97KK265 pKa = 10.23KK266 pKa = 9.73SSKK269 pKa = 10.33AGPSLEE275 pKa = 4.06LL276 pKa = 4.47
MM1 pKa = 7.52SNVKK5 pKa = 9.98QLTEE9 pKa = 4.08KK10 pKa = 10.51KK11 pKa = 10.05IKK13 pKa = 9.93EE14 pKa = 4.02LLAGGSADD22 pKa = 4.03VEE24 pKa = 4.45IEE26 pKa = 4.46TEE28 pKa = 4.09DD29 pKa = 3.57STPGFSFKK37 pKa = 10.96AFYY40 pKa = 8.24DD41 pKa = 3.67TNKK44 pKa = 10.16NLEE47 pKa = 4.03ITFTNCLNILKK58 pKa = 9.92CRR60 pKa = 11.84KK61 pKa = 9.63QIFAACKK68 pKa = 7.45SGKK71 pKa = 9.77YY72 pKa = 7.99VFCGKK77 pKa = 9.11TIVATNTDD85 pKa = 3.75VGPDD89 pKa = 2.87DD90 pKa = 3.44WTFKK94 pKa = 9.6RR95 pKa = 11.84TEE97 pKa = 3.6AFIRR101 pKa = 11.84TKK103 pKa = 9.04MASMVEE109 pKa = 3.99KK110 pKa = 10.61SKK112 pKa = 11.51NDD114 pKa = 3.46AAKK117 pKa = 10.37QEE119 pKa = 4.19MYY121 pKa = 10.86NKK123 pKa = 9.67IMEE126 pKa = 4.76LPLVAAYY133 pKa = 9.82GLNVPASFDD142 pKa = 3.51TCALRR147 pKa = 11.84MMLCIGGPLPLLSSMTGLAPIIFPLAYY174 pKa = 8.37YY175 pKa = 10.6QNVKK179 pKa = 10.31KK180 pKa = 10.66EE181 pKa = 3.8KK182 pKa = 10.6LGIKK186 pKa = 10.06NFSTYY191 pKa = 9.83EE192 pKa = 3.92QVCKK196 pKa = 9.96VAKK199 pKa = 10.09VLSASQIEE207 pKa = 4.69FKK209 pKa = 11.32NEE211 pKa = 3.41LEE213 pKa = 4.25EE214 pKa = 4.18MFKK217 pKa = 10.96SAVKK221 pKa = 10.36LLSEE225 pKa = 4.79SNPGTASSISLKK237 pKa = 10.73KK238 pKa = 10.13YY239 pKa = 10.29DD240 pKa = 3.9EE241 pKa = 4.12QVKK244 pKa = 10.65YY245 pKa = 9.75MDD247 pKa = 3.99KK248 pKa = 11.02AFSASLSMDD257 pKa = 3.49DD258 pKa = 4.54YY259 pKa = 11.95GEE261 pKa = 4.16HH262 pKa = 6.2SKK264 pKa = 10.97KK265 pKa = 10.23KK266 pKa = 9.73SSKK269 pKa = 10.33AGPSLEE275 pKa = 4.06LL276 pKa = 4.47
Molecular weight: 30.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5020 |
276 |
2877 |
1004.0 |
114.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.263 ± 0.499 | 2.271 ± 0.484 |
5.876 ± 0.423 | 6.394 ± 0.338 |
4.781 ± 0.254 | 4.084 ± 0.321 |
1.813 ± 0.247 | 8.227 ± 0.312 |
9.104 ± 0.395 | 9.163 ± 0.378 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.307 ± 0.551 | 6.315 ± 0.499 |
2.988 ± 0.277 | 2.968 ± 0.071 |
3.127 ± 0.28 | 9.542 ± 0.347 |
5.339 ± 0.211 | 5.598 ± 0.545 |
0.797 ± 0.174 | 4.044 ± 0.316 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
