
Phycomyces blakesleeanus (strain ATCC 8743b / DSM 1359 / FGSC 10004 / NBRC 33097 / NRRL 1555)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Mucoromycota; Mucoromycotina; Mucoromycetes; Mucorales; Phycomycetaceae; Phycomyces; Phycomyces blakesleeanus
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
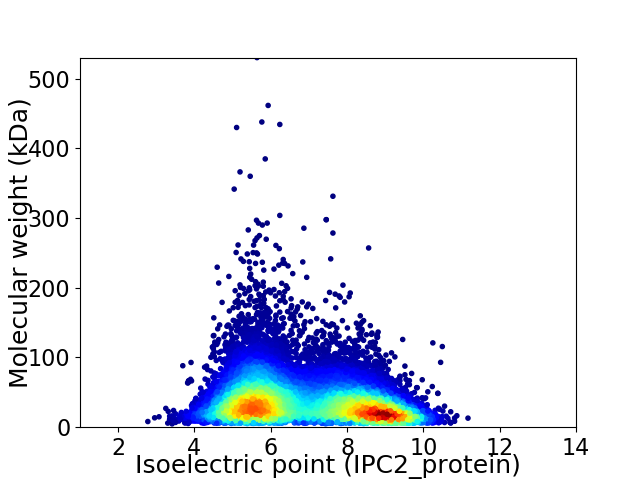
Virtual 2D-PAGE plot for 16155 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A162ZRP6|A0A162ZRP6_PHYB8 Transcription factor E2F/dimerization partner OS=Phycomyces blakesleeanus (strain ATCC 8743b / DSM 1359 / FGSC 10004 / NBRC 33097 / NRRL 1555) OX=763407 GN=PHYBLDRAFT_188617 PE=3 SV=1
MM1 pKa = 7.62LSYY4 pKa = 10.19PIQLALQTLCLASAFLCALLFAVISSLPVPSVLSMGTSSYY44 pKa = 11.06RR45 pKa = 11.84MRR47 pKa = 11.84IIHH50 pKa = 6.87KK51 pKa = 10.26DD52 pKa = 3.35SEE54 pKa = 4.51GCLQSFCTSSEE65 pKa = 4.11VFSSGFLRR73 pKa = 11.84CGSLGKK79 pKa = 8.42TLGSMSCPAVSSEE92 pKa = 3.97IKK94 pKa = 10.46SAGDD98 pKa = 3.27VEE100 pKa = 4.5GLSRR104 pKa = 11.84STSSNQQNSICHH116 pKa = 6.0RR117 pKa = 11.84MNSTVRR123 pKa = 11.84EE124 pKa = 4.13DD125 pKa = 4.07DD126 pKa = 3.34KK127 pKa = 11.05HH128 pKa = 7.65SHH130 pKa = 5.4MEE132 pKa = 3.76RR133 pKa = 11.84VAYY136 pKa = 9.8IPVRR140 pKa = 11.84HH141 pKa = 6.2EE142 pKa = 4.1NQGSKK147 pKa = 10.87CMVRR151 pKa = 11.84TRR153 pKa = 11.84LPYY156 pKa = 9.78YY157 pKa = 10.85GEE159 pKa = 4.36DD160 pKa = 3.4LEE162 pKa = 4.52EE163 pKa = 4.35QMFEE167 pKa = 4.13VLVSKK172 pKa = 8.75TSPLEE177 pKa = 4.18LIRR180 pKa = 11.84LASEE184 pKa = 4.17PLGSDD189 pKa = 3.35NQSEE193 pKa = 4.93PYY195 pKa = 10.18SSGYY199 pKa = 11.04DD200 pKa = 3.63EE201 pKa = 4.94VCVGAPAIEE210 pKa = 4.72CFLALPCPFEE220 pKa = 3.89QGTFNRR226 pKa = 11.84HH227 pKa = 5.25RR228 pKa = 11.84PSHH231 pKa = 6.7PLTDD235 pKa = 5.46EE236 pKa = 4.48IPSSGQNMILSDD248 pKa = 3.72ISDD251 pKa = 3.73LTSLLEE257 pKa = 4.37AMTISDD263 pKa = 3.65ASEE266 pKa = 4.25DD267 pKa = 3.52LSAGQNSFANTAQQVGGVGIEE288 pKa = 3.9VASAIEE294 pKa = 4.0TRR296 pKa = 11.84WYY298 pKa = 10.77SEE300 pKa = 3.57NAPWFEE306 pKa = 5.38EE307 pKa = 4.32IPQCSAFVGGFFAFVQEE324 pKa = 4.21EE325 pKa = 4.16PMDD328 pKa = 3.9FVSTFDD334 pKa = 3.9APLEE338 pKa = 3.98QMMTDD343 pKa = 3.31ADD345 pKa = 4.58EE346 pKa = 4.93EE347 pKa = 4.75DD348 pKa = 4.76FDD350 pKa = 6.25CSMSTISEE358 pKa = 4.44PEE360 pKa = 3.78TSEE363 pKa = 4.01MLDD366 pKa = 3.31VEE368 pKa = 4.25YY369 pKa = 11.09DD370 pKa = 3.97GIVVAGVSTACWYY383 pKa = 10.39NIHH386 pKa = 7.61DD387 pKa = 4.51EE388 pKa = 4.13LAMVPEE394 pKa = 4.62DD395 pKa = 3.65LCGDD399 pKa = 3.9SLMDD403 pKa = 3.95LAVEE407 pKa = 4.01VVPGIEE413 pKa = 5.15GILDD417 pKa = 4.13HH418 pKa = 7.05YY419 pKa = 11.29RR420 pKa = 11.84DD421 pKa = 4.21DD422 pKa = 5.13CLMYY426 pKa = 9.74WDD428 pKa = 4.95EE429 pKa = 4.37SLVPKK434 pKa = 10.68GSAIMMATMFPDD446 pKa = 3.18SGVAGSFPLDD456 pKa = 3.49QANMTNKK463 pKa = 10.18GATCVPQQVAEE474 pKa = 4.19DD475 pKa = 4.25LVWSPEE481 pKa = 3.78KK482 pKa = 10.42EE483 pKa = 3.96PRR485 pKa = 11.84VPSSPLLPGSDD496 pKa = 3.75SGADD500 pKa = 3.46LPGQSGVSLPEE511 pKa = 3.78VEE513 pKa = 5.19APEE516 pKa = 4.77PPTIPGVVVWLTTAPEE532 pKa = 4.32SPVKK536 pKa = 10.66VLEE539 pKa = 4.54EE540 pKa = 3.95EE541 pKa = 4.76DD542 pKa = 5.59KK543 pKa = 11.51EE544 pKa = 5.64DD545 pKa = 5.8DD546 pKa = 5.37DD547 pKa = 7.0DD548 pKa = 6.68DD549 pKa = 6.01EE550 pKa = 5.05EE551 pKa = 6.54DD552 pKa = 3.56SDD554 pKa = 5.24EE555 pKa = 4.95GYY557 pKa = 11.11EE558 pKa = 4.82DD559 pKa = 5.05DD560 pKa = 6.06DD561 pKa = 5.39EE562 pKa = 5.9GVTLQDD568 pKa = 3.4SALADD573 pKa = 3.77LLAEE577 pKa = 4.38LEE579 pKa = 4.32EE580 pKa = 5.15SDD582 pKa = 5.6FDD584 pKa = 5.35DD585 pKa = 4.39EE586 pKa = 4.67AA587 pKa = 6.32
MM1 pKa = 7.62LSYY4 pKa = 10.19PIQLALQTLCLASAFLCALLFAVISSLPVPSVLSMGTSSYY44 pKa = 11.06RR45 pKa = 11.84MRR47 pKa = 11.84IIHH50 pKa = 6.87KK51 pKa = 10.26DD52 pKa = 3.35SEE54 pKa = 4.51GCLQSFCTSSEE65 pKa = 4.11VFSSGFLRR73 pKa = 11.84CGSLGKK79 pKa = 8.42TLGSMSCPAVSSEE92 pKa = 3.97IKK94 pKa = 10.46SAGDD98 pKa = 3.27VEE100 pKa = 4.5GLSRR104 pKa = 11.84STSSNQQNSICHH116 pKa = 6.0RR117 pKa = 11.84MNSTVRR123 pKa = 11.84EE124 pKa = 4.13DD125 pKa = 4.07DD126 pKa = 3.34KK127 pKa = 11.05HH128 pKa = 7.65SHH130 pKa = 5.4MEE132 pKa = 3.76RR133 pKa = 11.84VAYY136 pKa = 9.8IPVRR140 pKa = 11.84HH141 pKa = 6.2EE142 pKa = 4.1NQGSKK147 pKa = 10.87CMVRR151 pKa = 11.84TRR153 pKa = 11.84LPYY156 pKa = 9.78YY157 pKa = 10.85GEE159 pKa = 4.36DD160 pKa = 3.4LEE162 pKa = 4.52EE163 pKa = 4.35QMFEE167 pKa = 4.13VLVSKK172 pKa = 8.75TSPLEE177 pKa = 4.18LIRR180 pKa = 11.84LASEE184 pKa = 4.17PLGSDD189 pKa = 3.35NQSEE193 pKa = 4.93PYY195 pKa = 10.18SSGYY199 pKa = 11.04DD200 pKa = 3.63EE201 pKa = 4.94VCVGAPAIEE210 pKa = 4.72CFLALPCPFEE220 pKa = 3.89QGTFNRR226 pKa = 11.84HH227 pKa = 5.25RR228 pKa = 11.84PSHH231 pKa = 6.7PLTDD235 pKa = 5.46EE236 pKa = 4.48IPSSGQNMILSDD248 pKa = 3.72ISDD251 pKa = 3.73LTSLLEE257 pKa = 4.37AMTISDD263 pKa = 3.65ASEE266 pKa = 4.25DD267 pKa = 3.52LSAGQNSFANTAQQVGGVGIEE288 pKa = 3.9VASAIEE294 pKa = 4.0TRR296 pKa = 11.84WYY298 pKa = 10.77SEE300 pKa = 3.57NAPWFEE306 pKa = 5.38EE307 pKa = 4.32IPQCSAFVGGFFAFVQEE324 pKa = 4.21EE325 pKa = 4.16PMDD328 pKa = 3.9FVSTFDD334 pKa = 3.9APLEE338 pKa = 3.98QMMTDD343 pKa = 3.31ADD345 pKa = 4.58EE346 pKa = 4.93EE347 pKa = 4.75DD348 pKa = 4.76FDD350 pKa = 6.25CSMSTISEE358 pKa = 4.44PEE360 pKa = 3.78TSEE363 pKa = 4.01MLDD366 pKa = 3.31VEE368 pKa = 4.25YY369 pKa = 11.09DD370 pKa = 3.97GIVVAGVSTACWYY383 pKa = 10.39NIHH386 pKa = 7.61DD387 pKa = 4.51EE388 pKa = 4.13LAMVPEE394 pKa = 4.62DD395 pKa = 3.65LCGDD399 pKa = 3.9SLMDD403 pKa = 3.95LAVEE407 pKa = 4.01VVPGIEE413 pKa = 5.15GILDD417 pKa = 4.13HH418 pKa = 7.05YY419 pKa = 11.29RR420 pKa = 11.84DD421 pKa = 4.21DD422 pKa = 5.13CLMYY426 pKa = 9.74WDD428 pKa = 4.95EE429 pKa = 4.37SLVPKK434 pKa = 10.68GSAIMMATMFPDD446 pKa = 3.18SGVAGSFPLDD456 pKa = 3.49QANMTNKK463 pKa = 10.18GATCVPQQVAEE474 pKa = 4.19DD475 pKa = 4.25LVWSPEE481 pKa = 3.78KK482 pKa = 10.42EE483 pKa = 3.96PRR485 pKa = 11.84VPSSPLLPGSDD496 pKa = 3.75SGADD500 pKa = 3.46LPGQSGVSLPEE511 pKa = 3.78VEE513 pKa = 5.19APEE516 pKa = 4.77PPTIPGVVVWLTTAPEE532 pKa = 4.32SPVKK536 pKa = 10.66VLEE539 pKa = 4.54EE540 pKa = 3.95EE541 pKa = 4.76DD542 pKa = 5.59KK543 pKa = 11.51EE544 pKa = 5.64DD545 pKa = 5.8DD546 pKa = 5.37DD547 pKa = 7.0DD548 pKa = 6.68DD549 pKa = 6.01EE550 pKa = 5.05EE551 pKa = 6.54DD552 pKa = 3.56SDD554 pKa = 5.24EE555 pKa = 4.95GYY557 pKa = 11.11EE558 pKa = 4.82DD559 pKa = 5.05DD560 pKa = 6.06DD561 pKa = 5.39EE562 pKa = 5.9GVTLQDD568 pKa = 3.4SALADD573 pKa = 3.77LLAEE577 pKa = 4.38LEE579 pKa = 4.32EE580 pKa = 5.15SDD582 pKa = 5.6FDD584 pKa = 5.35DD585 pKa = 4.39EE586 pKa = 4.67AA587 pKa = 6.32
Molecular weight: 63.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A162Y7C6|A0A162Y7C6_PHYB8 Uncharacterized protein OS=Phycomyces blakesleeanus (strain ATCC 8743b / DSM 1359 / FGSC 10004 / NBRC 33097 / NRRL 1555) OX=763407 GN=PHYBLDRAFT_140823 PE=4 SV=1
MM1 pKa = 8.02SMLRR5 pKa = 11.84QLWSSAVSRR14 pKa = 11.84LVPSTTNITQNTTISRR30 pKa = 11.84FTAARR35 pKa = 11.84PSMLGANTGSFLSSSFGLPRR55 pKa = 11.84SPFAASPFAATQMRR69 pKa = 11.84FVTYY73 pKa = 10.59GNTYY77 pKa = 9.82QPSNLVRR84 pKa = 11.84KK85 pKa = 9.15RR86 pKa = 11.84RR87 pKa = 11.84HH88 pKa = 5.25GFLSRR93 pKa = 11.84VATKK97 pKa = 10.31NGRR100 pKa = 11.84RR101 pKa = 11.84VLARR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84LKK109 pKa = 10.19GRR111 pKa = 11.84KK112 pKa = 8.68FLTHH116 pKa = 6.84
MM1 pKa = 8.02SMLRR5 pKa = 11.84QLWSSAVSRR14 pKa = 11.84LVPSTTNITQNTTISRR30 pKa = 11.84FTAARR35 pKa = 11.84PSMLGANTGSFLSSSFGLPRR55 pKa = 11.84SPFAASPFAATQMRR69 pKa = 11.84FVTYY73 pKa = 10.59GNTYY77 pKa = 9.82QPSNLVRR84 pKa = 11.84KK85 pKa = 9.15RR86 pKa = 11.84RR87 pKa = 11.84HH88 pKa = 5.25GFLSRR93 pKa = 11.84VATKK97 pKa = 10.31NGRR100 pKa = 11.84RR101 pKa = 11.84VLARR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84LKK109 pKa = 10.19GRR111 pKa = 11.84KK112 pKa = 8.68FLTHH116 pKa = 6.84
Molecular weight: 12.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5761222 |
49 |
4633 |
356.6 |
40.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.434 ± 0.018 | 1.723 ± 0.009 |
5.426 ± 0.012 | 5.948 ± 0.017 |
4.253 ± 0.015 | 5.138 ± 0.018 |
2.701 ± 0.01 | 6.162 ± 0.015 |
6.085 ± 0.019 | 9.404 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.463 ± 0.008 | 4.837 ± 0.013 |
4.905 ± 0.016 | 4.16 ± 0.015 |
5.125 ± 0.014 | 8.339 ± 0.02 |
6.132 ± 0.014 | 6.137 ± 0.016 |
1.245 ± 0.006 | 3.382 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
