
Bromus-associated circular DNA virus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus bromas1; Bromus associated gemycircularvirus 1
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
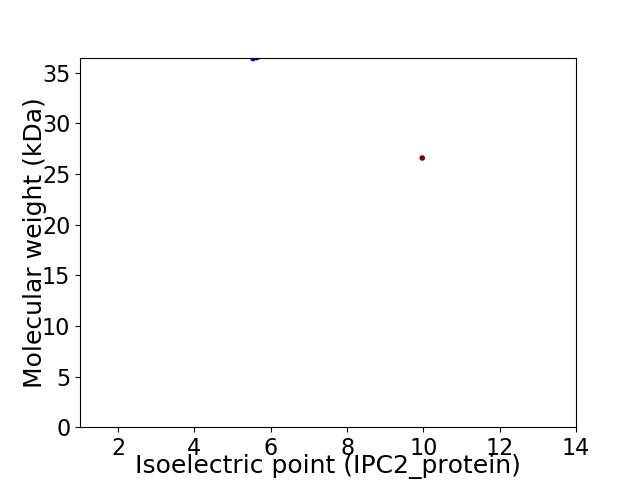
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4U9K5|A0A0B4U9K5_9VIRU Capsid protein OS=Bromus-associated circular DNA virus 3 OX=1590156 PE=4 SV=1
MM1 pKa = 7.87PSFDD5 pKa = 3.51LHH7 pKa = 6.63CRR9 pKa = 11.84YY10 pKa = 10.41ALLTYY15 pKa = 8.75AQCGEE20 pKa = 4.33LSADD24 pKa = 3.32AVGEE28 pKa = 3.91FLQGMGFSCVVGRR41 pKa = 11.84EE42 pKa = 3.95DD43 pKa = 3.46HH44 pKa = 6.6QDD46 pKa = 3.36GGVHH50 pKa = 5.79LHH52 pKa = 6.49CFVDD56 pKa = 4.74FGRR59 pKa = 11.84KK60 pKa = 7.8RR61 pKa = 11.84RR62 pKa = 11.84FRR64 pKa = 11.84RR65 pKa = 11.84VDD67 pKa = 3.21VFDD70 pKa = 3.69VEE72 pKa = 4.89GRR74 pKa = 11.84HH75 pKa = 6.09PNISPSKK82 pKa = 7.94GTPAKK87 pKa = 10.51GYY89 pKa = 10.65DD90 pKa = 3.43YY91 pKa = 10.68AIKK94 pKa = 10.32DD95 pKa = 3.5GRR97 pKa = 11.84IEE99 pKa = 4.17YY100 pKa = 10.42CSLDD104 pKa = 3.44RR105 pKa = 11.84PGPAEE110 pKa = 3.75KK111 pKa = 10.57SGTIEE116 pKa = 3.39KK117 pKa = 9.85WSRR120 pKa = 11.84ITSASDD126 pKa = 3.68RR127 pKa = 11.84EE128 pKa = 4.58SFWDD132 pKa = 4.13LVHH135 pKa = 7.0EE136 pKa = 5.17LDD138 pKa = 4.48PKK140 pKa = 10.85SAACCHH146 pKa = 5.82TQLQKK151 pKa = 10.73YY152 pKa = 8.84CDD154 pKa = 3.27WKK156 pKa = 10.89FAVIPAKK163 pKa = 10.52YY164 pKa = 8.54EE165 pKa = 3.92HH166 pKa = 7.29PPGITFFGGDD176 pKa = 3.58LDD178 pKa = 6.04GRR180 pKa = 11.84DD181 pKa = 3.79DD182 pKa = 3.7WLQQSGIGLGEE193 pKa = 4.2PLLGICLSICVYY205 pKa = 10.84GEE207 pKa = 3.84SRR209 pKa = 11.84TGKK212 pKa = 7.35TLWARR217 pKa = 11.84SLGSHH222 pKa = 7.11IYY224 pKa = 10.44CVGLVSGTEE233 pKa = 3.97CMKK236 pKa = 10.36ATEE239 pKa = 3.79ADD241 pKa = 3.68YY242 pKa = 11.48AVFDD246 pKa = 5.44DD247 pKa = 3.7IRR249 pKa = 11.84GGIKK253 pKa = 9.99FFPSFKK259 pKa = 10.33EE260 pKa = 3.72WLGCQAYY267 pKa = 7.85VTVKK271 pKa = 10.25CLYY274 pKa = 10.23RR275 pKa = 11.84EE276 pKa = 4.0PSLVKK281 pKa = 9.6WGKK284 pKa = 9.21PSIWLSNTDD293 pKa = 3.9PRR295 pKa = 11.84DD296 pKa = 3.34EE297 pKa = 4.42MLNADD302 pKa = 4.4IEE304 pKa = 4.54WMNKK308 pKa = 7.88NCIFVEE314 pKa = 4.28VNSAIFHH321 pKa = 6.72ANTT324 pKa = 3.54
MM1 pKa = 7.87PSFDD5 pKa = 3.51LHH7 pKa = 6.63CRR9 pKa = 11.84YY10 pKa = 10.41ALLTYY15 pKa = 8.75AQCGEE20 pKa = 4.33LSADD24 pKa = 3.32AVGEE28 pKa = 3.91FLQGMGFSCVVGRR41 pKa = 11.84EE42 pKa = 3.95DD43 pKa = 3.46HH44 pKa = 6.6QDD46 pKa = 3.36GGVHH50 pKa = 5.79LHH52 pKa = 6.49CFVDD56 pKa = 4.74FGRR59 pKa = 11.84KK60 pKa = 7.8RR61 pKa = 11.84RR62 pKa = 11.84FRR64 pKa = 11.84RR65 pKa = 11.84VDD67 pKa = 3.21VFDD70 pKa = 3.69VEE72 pKa = 4.89GRR74 pKa = 11.84HH75 pKa = 6.09PNISPSKK82 pKa = 7.94GTPAKK87 pKa = 10.51GYY89 pKa = 10.65DD90 pKa = 3.43YY91 pKa = 10.68AIKK94 pKa = 10.32DD95 pKa = 3.5GRR97 pKa = 11.84IEE99 pKa = 4.17YY100 pKa = 10.42CSLDD104 pKa = 3.44RR105 pKa = 11.84PGPAEE110 pKa = 3.75KK111 pKa = 10.57SGTIEE116 pKa = 3.39KK117 pKa = 9.85WSRR120 pKa = 11.84ITSASDD126 pKa = 3.68RR127 pKa = 11.84EE128 pKa = 4.58SFWDD132 pKa = 4.13LVHH135 pKa = 7.0EE136 pKa = 5.17LDD138 pKa = 4.48PKK140 pKa = 10.85SAACCHH146 pKa = 5.82TQLQKK151 pKa = 10.73YY152 pKa = 8.84CDD154 pKa = 3.27WKK156 pKa = 10.89FAVIPAKK163 pKa = 10.52YY164 pKa = 8.54EE165 pKa = 3.92HH166 pKa = 7.29PPGITFFGGDD176 pKa = 3.58LDD178 pKa = 6.04GRR180 pKa = 11.84DD181 pKa = 3.79DD182 pKa = 3.7WLQQSGIGLGEE193 pKa = 4.2PLLGICLSICVYY205 pKa = 10.84GEE207 pKa = 3.84SRR209 pKa = 11.84TGKK212 pKa = 7.35TLWARR217 pKa = 11.84SLGSHH222 pKa = 7.11IYY224 pKa = 10.44CVGLVSGTEE233 pKa = 3.97CMKK236 pKa = 10.36ATEE239 pKa = 3.79ADD241 pKa = 3.68YY242 pKa = 11.48AVFDD246 pKa = 5.44DD247 pKa = 3.7IRR249 pKa = 11.84GGIKK253 pKa = 9.99FFPSFKK259 pKa = 10.33EE260 pKa = 3.72WLGCQAYY267 pKa = 7.85VTVKK271 pKa = 10.25CLYY274 pKa = 10.23RR275 pKa = 11.84EE276 pKa = 4.0PSLVKK281 pKa = 9.6WGKK284 pKa = 9.21PSIWLSNTDD293 pKa = 3.9PRR295 pKa = 11.84DD296 pKa = 3.34EE297 pKa = 4.42MLNADD302 pKa = 4.4IEE304 pKa = 4.54WMNKK308 pKa = 7.88NCIFVEE314 pKa = 4.28VNSAIFHH321 pKa = 6.72ANTT324 pKa = 3.54
Molecular weight: 36.37 kDa
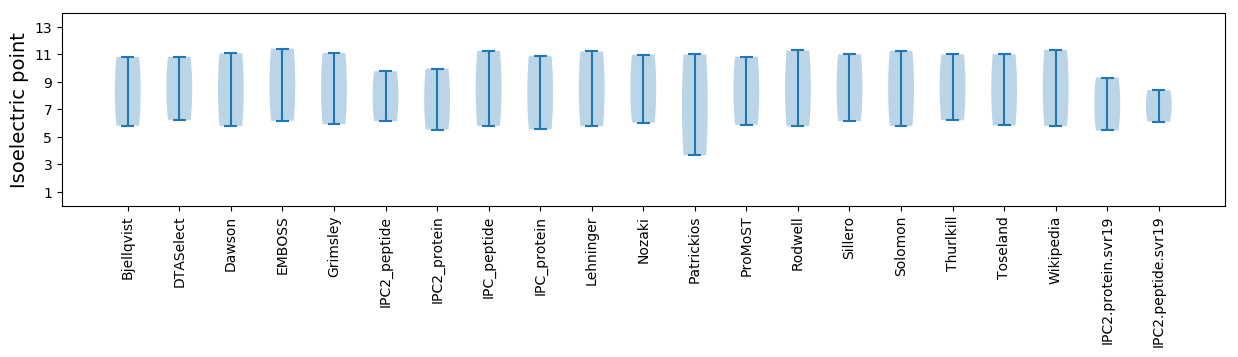
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4U9K5|A0A0B4U9K5_9VIRU Capsid protein OS=Bromus-associated circular DNA virus 3 OX=1590156 PE=4 SV=1
MM1 pKa = 7.92RR2 pKa = 11.84SRR4 pKa = 11.84YY5 pKa = 6.94TKK7 pKa = 9.83RR8 pKa = 11.84SGFRR12 pKa = 11.84RR13 pKa = 11.84TKK15 pKa = 7.74RR16 pKa = 11.84TSRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84SYY23 pKa = 10.79KK24 pKa = 9.86GRR26 pKa = 11.84TRR28 pKa = 11.84YY29 pKa = 7.36STRR32 pKa = 11.84RR33 pKa = 11.84VTKK36 pKa = 10.21RR37 pKa = 11.84LPQRR41 pKa = 11.84KK42 pKa = 7.61SRR44 pKa = 11.84KK45 pKa = 8.83MILNLTSKK53 pKa = 10.52KK54 pKa = 10.32KK55 pKa = 10.13RR56 pKa = 11.84DD57 pKa = 3.34TMAPNTGYY65 pKa = 9.33PAATNQIRR73 pKa = 11.84GLEE76 pKa = 4.03IQGGNVATITLWSPTARR93 pKa = 11.84EE94 pKa = 3.6LRR96 pKa = 11.84EE97 pKa = 3.59IFEE100 pKa = 4.49NDD102 pKa = 3.16KK103 pKa = 11.43SSVPSGRR110 pKa = 11.84EE111 pKa = 3.51ATNCYY116 pKa = 9.85IVGLKK121 pKa = 10.16EE122 pKa = 4.41KK123 pKa = 10.57ISILTNSGNPWKK135 pKa = 9.74WRR137 pKa = 11.84RR138 pKa = 11.84IAFTHH143 pKa = 6.85KK144 pKa = 10.49GLLPLGEE151 pKa = 4.14QFEE154 pKa = 4.56GSRR157 pKa = 11.84VYY159 pKa = 10.08STITDD164 pKa = 3.41VVGRR168 pKa = 11.84ITYY171 pKa = 9.95YY172 pKa = 9.51RR173 pKa = 11.84TLTPLPDD180 pKa = 3.47TSKK183 pKa = 9.96TPLVDD188 pKa = 3.81YY189 pKa = 10.28LFKK192 pKa = 11.12GQGEE196 pKa = 4.63GGLGVQDD203 pKa = 3.17WTDD206 pKa = 3.48IITAPVDD213 pKa = 3.31TTRR216 pKa = 11.84IKK218 pKa = 10.95VMHH221 pKa = 7.08DD222 pKa = 2.98HH223 pKa = 6.41THH225 pKa = 6.6PHH227 pKa = 6.55PKK229 pKa = 9.04WKK231 pKa = 10.45
MM1 pKa = 7.92RR2 pKa = 11.84SRR4 pKa = 11.84YY5 pKa = 6.94TKK7 pKa = 9.83RR8 pKa = 11.84SGFRR12 pKa = 11.84RR13 pKa = 11.84TKK15 pKa = 7.74RR16 pKa = 11.84TSRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84SYY23 pKa = 10.79KK24 pKa = 9.86GRR26 pKa = 11.84TRR28 pKa = 11.84YY29 pKa = 7.36STRR32 pKa = 11.84RR33 pKa = 11.84VTKK36 pKa = 10.21RR37 pKa = 11.84LPQRR41 pKa = 11.84KK42 pKa = 7.61SRR44 pKa = 11.84KK45 pKa = 8.83MILNLTSKK53 pKa = 10.52KK54 pKa = 10.32KK55 pKa = 10.13RR56 pKa = 11.84DD57 pKa = 3.34TMAPNTGYY65 pKa = 9.33PAATNQIRR73 pKa = 11.84GLEE76 pKa = 4.03IQGGNVATITLWSPTARR93 pKa = 11.84EE94 pKa = 3.6LRR96 pKa = 11.84EE97 pKa = 3.59IFEE100 pKa = 4.49NDD102 pKa = 3.16KK103 pKa = 11.43SSVPSGRR110 pKa = 11.84EE111 pKa = 3.51ATNCYY116 pKa = 9.85IVGLKK121 pKa = 10.16EE122 pKa = 4.41KK123 pKa = 10.57ISILTNSGNPWKK135 pKa = 9.74WRR137 pKa = 11.84RR138 pKa = 11.84IAFTHH143 pKa = 6.85KK144 pKa = 10.49GLLPLGEE151 pKa = 4.14QFEE154 pKa = 4.56GSRR157 pKa = 11.84VYY159 pKa = 10.08STITDD164 pKa = 3.41VVGRR168 pKa = 11.84ITYY171 pKa = 9.95YY172 pKa = 9.51RR173 pKa = 11.84TLTPLPDD180 pKa = 3.47TSKK183 pKa = 9.96TPLVDD188 pKa = 3.81YY189 pKa = 10.28LFKK192 pKa = 11.12GQGEE196 pKa = 4.63GGLGVQDD203 pKa = 3.17WTDD206 pKa = 3.48IITAPVDD213 pKa = 3.31TTRR216 pKa = 11.84IKK218 pKa = 10.95VMHH221 pKa = 7.08DD222 pKa = 2.98HH223 pKa = 6.41THH225 pKa = 6.6PHH227 pKa = 6.55PKK229 pKa = 9.04WKK231 pKa = 10.45
Molecular weight: 26.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
555 |
231 |
324 |
277.5 |
31.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.045 ± 0.971 | 2.883 ± 1.504 |
5.946 ± 1.259 | 5.045 ± 0.706 |
3.964 ± 1.105 | 8.829 ± 0.637 |
2.703 ± 0.33 | 5.586 ± 0.292 |
6.667 ± 0.957 | 7.207 ± 0.172 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.622 ± 0.068 | 2.703 ± 0.467 |
5.045 ± 0.358 | 2.523 ± 0.046 |
8.108 ± 2.198 | 7.027 ± 0.062 |
7.387 ± 2.907 | 5.405 ± 0.395 |
2.523 ± 0.22 | 3.784 ± 0.069 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
