
Pseudooceanicola batsensis (strain ATCC BAA-863 / DSM 15984 / KCTC 12145 / HTCC2597) (Oceanicola batsensis)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pseudooceanicola; Pseudooceanicola batsensis
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
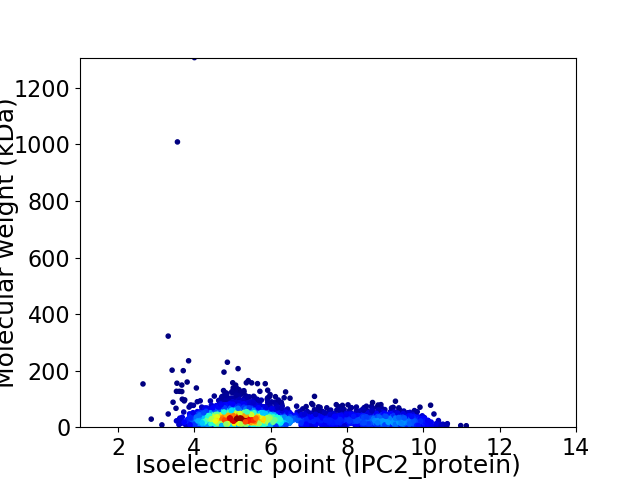
Virtual 2D-PAGE plot for 4193 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3U2A6|A3U2A6_PSEBH Uncharacterized protein OS=Pseudooceanicola batsensis (strain ATCC BAA-863 / DSM 15984 / KCTC 12145 / HTCC2597) OX=252305 GN=OB2597_14721 PE=4 SV=1
MM1 pKa = 7.28TYY3 pKa = 10.3YY4 pKa = 10.25YY5 pKa = 10.86SKK7 pKa = 9.73YY8 pKa = 10.4SYY10 pKa = 10.36YY11 pKa = 10.69NKK13 pKa = 10.02HH14 pKa = 6.22SYY16 pKa = 9.9SEE18 pKa = 4.67KK19 pKa = 9.5KK20 pKa = 10.56YY21 pKa = 9.55EE22 pKa = 3.68WTAYY26 pKa = 9.56TEE28 pKa = 4.24ADD30 pKa = 4.17LLGDD34 pKa = 4.14HH35 pKa = 6.7NGKK38 pKa = 10.62SIDD41 pKa = 4.1CGDD44 pKa = 4.1TFPMPGSATVCMSTYY59 pKa = 11.24DD60 pKa = 3.41NDD62 pKa = 4.46GYY64 pKa = 11.58LSGDD68 pKa = 3.77SSWWSDD74 pKa = 3.98DD75 pKa = 3.22KK76 pKa = 11.58ATDD79 pKa = 3.49STGQKK84 pKa = 10.64AYY86 pKa = 9.89IDD88 pKa = 3.86GGRR91 pKa = 11.84EE92 pKa = 3.85GGQLYY97 pKa = 10.59AEE99 pKa = 4.51QYY101 pKa = 10.17HH102 pKa = 5.26VLKK105 pKa = 10.99GSDD108 pKa = 2.72GKK110 pKa = 11.04YY111 pKa = 9.66YY112 pKa = 10.4YY113 pKa = 10.48LIEE116 pKa = 5.39IEE118 pKa = 4.3VEE120 pKa = 4.51GYY122 pKa = 8.91DD123 pKa = 4.73APGKK127 pKa = 10.39GDD129 pKa = 4.38DD130 pKa = 3.63YY131 pKa = 10.77FTFYY135 pKa = 11.33GKK137 pKa = 10.21VPPAGVEE144 pKa = 4.07LEE146 pKa = 4.41VVSTKK151 pKa = 10.66NVWGCMIDD159 pKa = 3.83YY160 pKa = 9.76RR161 pKa = 11.84CLGAGDD167 pKa = 4.18KK168 pKa = 10.97APEE171 pKa = 3.92NTPPEE176 pKa = 4.22FTNDD180 pKa = 3.06PKK182 pKa = 11.08YY183 pKa = 11.24GKK185 pKa = 9.6ICIDD189 pKa = 3.57EE190 pKa = 4.36NSTFVIDD197 pKa = 5.92LNAKK201 pKa = 10.32DD202 pKa = 5.14DD203 pKa = 4.9DD204 pKa = 5.09GDD206 pKa = 3.77TLTYY210 pKa = 10.19EE211 pKa = 4.12IVGGEE216 pKa = 4.04DD217 pKa = 3.24ADD219 pKa = 3.8KK220 pKa = 11.4FEE222 pKa = 5.83IDD224 pKa = 3.76AEE226 pKa = 4.31TGEE229 pKa = 5.15LRR231 pKa = 11.84FKK233 pKa = 10.7DD234 pKa = 3.5APDD237 pKa = 3.61YY238 pKa = 10.46EE239 pKa = 4.33NPTDD243 pKa = 3.58KK244 pKa = 11.67GEE246 pKa = 4.49DD247 pKa = 2.99NVYY250 pKa = 9.43EE251 pKa = 4.36VKK253 pKa = 11.08VKK255 pKa = 10.93VSDD258 pKa = 3.91GNGGEE263 pKa = 4.35DD264 pKa = 3.28TTKK267 pKa = 10.99LKK269 pKa = 11.1VCVEE273 pKa = 4.21DD274 pKa = 5.08VEE276 pKa = 4.48EE277 pKa = 4.05QTGKK281 pKa = 10.79CIVIEE286 pKa = 4.29AEE288 pKa = 5.56DD289 pKa = 3.47MCEE292 pKa = 3.73WGFSKK297 pKa = 10.54AWGCDD302 pKa = 3.01ASGGKK307 pKa = 8.94LVKK310 pKa = 10.36LNCAGGQGKK319 pKa = 9.15LWTTFDD325 pKa = 4.32GPSGTYY331 pKa = 9.97DD332 pKa = 3.39FKK334 pKa = 11.13IYY336 pKa = 10.67AQDD339 pKa = 3.66EE340 pKa = 4.26NDD342 pKa = 3.94GQSKK346 pKa = 10.34LKK348 pKa = 10.35IYY350 pKa = 10.64INGEE354 pKa = 3.55FKK356 pKa = 10.83GYY358 pKa = 10.25IKK360 pKa = 10.66LDD362 pKa = 3.75RR363 pKa = 11.84DD364 pKa = 3.6SDD366 pKa = 4.2GSGSNHH372 pKa = 5.05GTFSTFVLEE381 pKa = 5.51DD382 pKa = 3.22IEE384 pKa = 4.45INKK387 pKa = 10.01GDD389 pKa = 4.04TIKK392 pKa = 10.39IYY394 pKa = 11.12ADD396 pKa = 3.73GDD398 pKa = 3.81CNEE401 pKa = 4.25YY402 pKa = 10.74VRR404 pKa = 11.84IDD406 pKa = 3.95KK407 pKa = 10.79IEE409 pKa = 3.96LCKK412 pKa = 10.39DD413 pKa = 3.68DD414 pKa = 5.18KK415 pKa = 11.23PLGAIGDD422 pKa = 4.4TVWYY426 pKa = 8.3DD427 pKa = 3.24TDD429 pKa = 4.31RR430 pKa = 11.84DD431 pKa = 4.06GVQDD435 pKa = 3.98DD436 pKa = 4.67GEE438 pKa = 5.62AGVEE442 pKa = 3.96NVTVNLLNGAGVVIATQMTDD462 pKa = 2.77VNGNYY467 pKa = 10.19LFEE470 pKa = 4.29EE471 pKa = 4.91LEE473 pKa = 4.18AGDD476 pKa = 3.71YY477 pKa = 10.39KK478 pKa = 11.41VEE480 pKa = 4.35FEE482 pKa = 4.79LPDD485 pKa = 3.65GNFVFTTQDD494 pKa = 2.99AGSDD498 pKa = 3.69DD499 pKa = 5.24AVDD502 pKa = 3.86SDD504 pKa = 4.87ADD506 pKa = 3.95TTTGLTDD513 pKa = 4.08TIALGEE519 pKa = 4.5GEE521 pKa = 4.53TNLTVDD527 pKa = 4.42AGLYY531 pKa = 10.27DD532 pKa = 4.65PNDD535 pKa = 3.82HH536 pKa = 7.55PEE538 pKa = 4.23PTDD541 pKa = 4.26DD542 pKa = 5.03AAKK545 pKa = 9.4TCADD549 pKa = 3.59EE550 pKa = 4.55EE551 pKa = 4.54KK552 pKa = 10.28TVDD555 pKa = 3.62VLANDD560 pKa = 3.87SDD562 pKa = 4.66PNGDD566 pKa = 3.56TLTITHH572 pKa = 6.58VDD574 pKa = 3.41GQAITEE580 pKa = 4.31GGEE583 pKa = 4.21VTTSAGTLVKK593 pKa = 10.61LLGGMLVVDD602 pKa = 5.04GEE604 pKa = 4.36DD605 pKa = 3.26AYY607 pKa = 10.77EE608 pKa = 4.38DD609 pKa = 4.07LDD611 pKa = 3.87IAEE614 pKa = 5.08HH615 pKa = 6.49AMEE618 pKa = 5.73DD619 pKa = 2.92ISYY622 pKa = 8.82TVSDD626 pKa = 4.02GMGGSATANLEE637 pKa = 3.95MTFCGDD643 pKa = 3.23ANTVEE648 pKa = 4.56SFVGYY653 pKa = 9.12MPEE656 pKa = 4.01TVSYY660 pKa = 10.49KK661 pKa = 10.56VADD664 pKa = 4.0GLRR667 pKa = 11.84NDD669 pKa = 3.84PYY671 pKa = 11.27EE672 pKa = 4.36DD673 pKa = 2.95VGYY676 pKa = 10.43DD677 pKa = 3.36ILITEE682 pKa = 4.42SDD684 pKa = 3.34NGRR687 pKa = 11.84LDD689 pKa = 3.61GVEE692 pKa = 4.57FTAAYY697 pKa = 10.25CLDD700 pKa = 3.88FNRR703 pKa = 11.84SITTGADD710 pKa = 3.02YY711 pKa = 11.21DD712 pKa = 4.06SAAEE716 pKa = 3.98LDD718 pKa = 3.11GDD720 pKa = 3.59MFVFNDD726 pKa = 3.62DD727 pKa = 3.57TASSVLLASQNGVNGAALDD746 pKa = 3.94NVNEE750 pKa = 4.4INWILNQDD758 pKa = 3.9FEE760 pKa = 5.59SDD762 pKa = 3.41GTGSVDD768 pKa = 3.33GKK770 pKa = 10.28FSGWEE775 pKa = 3.52VQYY778 pKa = 10.94AIWEE782 pKa = 4.26LTNNFPSDD790 pKa = 3.29AALNATPSNGQVEE803 pKa = 4.61DD804 pKa = 2.8VDD806 pKa = 4.88YY807 pKa = 10.75IVSRR811 pKa = 11.84AISEE815 pKa = 4.66GYY817 pKa = 10.39DD818 pKa = 3.35FEE820 pKa = 7.23AGVGDD825 pKa = 4.54IIGLVITPDD834 pKa = 3.23PVTAQNAQPFLVGVEE849 pKa = 4.09FEE851 pKa = 5.22TYY853 pKa = 10.71DD854 pKa = 3.8CLCC857 pKa = 4.56
MM1 pKa = 7.28TYY3 pKa = 10.3YY4 pKa = 10.25YY5 pKa = 10.86SKK7 pKa = 9.73YY8 pKa = 10.4SYY10 pKa = 10.36YY11 pKa = 10.69NKK13 pKa = 10.02HH14 pKa = 6.22SYY16 pKa = 9.9SEE18 pKa = 4.67KK19 pKa = 9.5KK20 pKa = 10.56YY21 pKa = 9.55EE22 pKa = 3.68WTAYY26 pKa = 9.56TEE28 pKa = 4.24ADD30 pKa = 4.17LLGDD34 pKa = 4.14HH35 pKa = 6.7NGKK38 pKa = 10.62SIDD41 pKa = 4.1CGDD44 pKa = 4.1TFPMPGSATVCMSTYY59 pKa = 11.24DD60 pKa = 3.41NDD62 pKa = 4.46GYY64 pKa = 11.58LSGDD68 pKa = 3.77SSWWSDD74 pKa = 3.98DD75 pKa = 3.22KK76 pKa = 11.58ATDD79 pKa = 3.49STGQKK84 pKa = 10.64AYY86 pKa = 9.89IDD88 pKa = 3.86GGRR91 pKa = 11.84EE92 pKa = 3.85GGQLYY97 pKa = 10.59AEE99 pKa = 4.51QYY101 pKa = 10.17HH102 pKa = 5.26VLKK105 pKa = 10.99GSDD108 pKa = 2.72GKK110 pKa = 11.04YY111 pKa = 9.66YY112 pKa = 10.4YY113 pKa = 10.48LIEE116 pKa = 5.39IEE118 pKa = 4.3VEE120 pKa = 4.51GYY122 pKa = 8.91DD123 pKa = 4.73APGKK127 pKa = 10.39GDD129 pKa = 4.38DD130 pKa = 3.63YY131 pKa = 10.77FTFYY135 pKa = 11.33GKK137 pKa = 10.21VPPAGVEE144 pKa = 4.07LEE146 pKa = 4.41VVSTKK151 pKa = 10.66NVWGCMIDD159 pKa = 3.83YY160 pKa = 9.76RR161 pKa = 11.84CLGAGDD167 pKa = 4.18KK168 pKa = 10.97APEE171 pKa = 3.92NTPPEE176 pKa = 4.22FTNDD180 pKa = 3.06PKK182 pKa = 11.08YY183 pKa = 11.24GKK185 pKa = 9.6ICIDD189 pKa = 3.57EE190 pKa = 4.36NSTFVIDD197 pKa = 5.92LNAKK201 pKa = 10.32DD202 pKa = 5.14DD203 pKa = 4.9DD204 pKa = 5.09GDD206 pKa = 3.77TLTYY210 pKa = 10.19EE211 pKa = 4.12IVGGEE216 pKa = 4.04DD217 pKa = 3.24ADD219 pKa = 3.8KK220 pKa = 11.4FEE222 pKa = 5.83IDD224 pKa = 3.76AEE226 pKa = 4.31TGEE229 pKa = 5.15LRR231 pKa = 11.84FKK233 pKa = 10.7DD234 pKa = 3.5APDD237 pKa = 3.61YY238 pKa = 10.46EE239 pKa = 4.33NPTDD243 pKa = 3.58KK244 pKa = 11.67GEE246 pKa = 4.49DD247 pKa = 2.99NVYY250 pKa = 9.43EE251 pKa = 4.36VKK253 pKa = 11.08VKK255 pKa = 10.93VSDD258 pKa = 3.91GNGGEE263 pKa = 4.35DD264 pKa = 3.28TTKK267 pKa = 10.99LKK269 pKa = 11.1VCVEE273 pKa = 4.21DD274 pKa = 5.08VEE276 pKa = 4.48EE277 pKa = 4.05QTGKK281 pKa = 10.79CIVIEE286 pKa = 4.29AEE288 pKa = 5.56DD289 pKa = 3.47MCEE292 pKa = 3.73WGFSKK297 pKa = 10.54AWGCDD302 pKa = 3.01ASGGKK307 pKa = 8.94LVKK310 pKa = 10.36LNCAGGQGKK319 pKa = 9.15LWTTFDD325 pKa = 4.32GPSGTYY331 pKa = 9.97DD332 pKa = 3.39FKK334 pKa = 11.13IYY336 pKa = 10.67AQDD339 pKa = 3.66EE340 pKa = 4.26NDD342 pKa = 3.94GQSKK346 pKa = 10.34LKK348 pKa = 10.35IYY350 pKa = 10.64INGEE354 pKa = 3.55FKK356 pKa = 10.83GYY358 pKa = 10.25IKK360 pKa = 10.66LDD362 pKa = 3.75RR363 pKa = 11.84DD364 pKa = 3.6SDD366 pKa = 4.2GSGSNHH372 pKa = 5.05GTFSTFVLEE381 pKa = 5.51DD382 pKa = 3.22IEE384 pKa = 4.45INKK387 pKa = 10.01GDD389 pKa = 4.04TIKK392 pKa = 10.39IYY394 pKa = 11.12ADD396 pKa = 3.73GDD398 pKa = 3.81CNEE401 pKa = 4.25YY402 pKa = 10.74VRR404 pKa = 11.84IDD406 pKa = 3.95KK407 pKa = 10.79IEE409 pKa = 3.96LCKK412 pKa = 10.39DD413 pKa = 3.68DD414 pKa = 5.18KK415 pKa = 11.23PLGAIGDD422 pKa = 4.4TVWYY426 pKa = 8.3DD427 pKa = 3.24TDD429 pKa = 4.31RR430 pKa = 11.84DD431 pKa = 4.06GVQDD435 pKa = 3.98DD436 pKa = 4.67GEE438 pKa = 5.62AGVEE442 pKa = 3.96NVTVNLLNGAGVVIATQMTDD462 pKa = 2.77VNGNYY467 pKa = 10.19LFEE470 pKa = 4.29EE471 pKa = 4.91LEE473 pKa = 4.18AGDD476 pKa = 3.71YY477 pKa = 10.39KK478 pKa = 11.41VEE480 pKa = 4.35FEE482 pKa = 4.79LPDD485 pKa = 3.65GNFVFTTQDD494 pKa = 2.99AGSDD498 pKa = 3.69DD499 pKa = 5.24AVDD502 pKa = 3.86SDD504 pKa = 4.87ADD506 pKa = 3.95TTTGLTDD513 pKa = 4.08TIALGEE519 pKa = 4.5GEE521 pKa = 4.53TNLTVDD527 pKa = 4.42AGLYY531 pKa = 10.27DD532 pKa = 4.65PNDD535 pKa = 3.82HH536 pKa = 7.55PEE538 pKa = 4.23PTDD541 pKa = 4.26DD542 pKa = 5.03AAKK545 pKa = 9.4TCADD549 pKa = 3.59EE550 pKa = 4.55EE551 pKa = 4.54KK552 pKa = 10.28TVDD555 pKa = 3.62VLANDD560 pKa = 3.87SDD562 pKa = 4.66PNGDD566 pKa = 3.56TLTITHH572 pKa = 6.58VDD574 pKa = 3.41GQAITEE580 pKa = 4.31GGEE583 pKa = 4.21VTTSAGTLVKK593 pKa = 10.61LLGGMLVVDD602 pKa = 5.04GEE604 pKa = 4.36DD605 pKa = 3.26AYY607 pKa = 10.77EE608 pKa = 4.38DD609 pKa = 4.07LDD611 pKa = 3.87IAEE614 pKa = 5.08HH615 pKa = 6.49AMEE618 pKa = 5.73DD619 pKa = 2.92ISYY622 pKa = 8.82TVSDD626 pKa = 4.02GMGGSATANLEE637 pKa = 3.95MTFCGDD643 pKa = 3.23ANTVEE648 pKa = 4.56SFVGYY653 pKa = 9.12MPEE656 pKa = 4.01TVSYY660 pKa = 10.49KK661 pKa = 10.56VADD664 pKa = 4.0GLRR667 pKa = 11.84NDD669 pKa = 3.84PYY671 pKa = 11.27EE672 pKa = 4.36DD673 pKa = 2.95VGYY676 pKa = 10.43DD677 pKa = 3.36ILITEE682 pKa = 4.42SDD684 pKa = 3.34NGRR687 pKa = 11.84LDD689 pKa = 3.61GVEE692 pKa = 4.57FTAAYY697 pKa = 10.25CLDD700 pKa = 3.88FNRR703 pKa = 11.84SITTGADD710 pKa = 3.02YY711 pKa = 11.21DD712 pKa = 4.06SAAEE716 pKa = 3.98LDD718 pKa = 3.11GDD720 pKa = 3.59MFVFNDD726 pKa = 3.62DD727 pKa = 3.57TASSVLLASQNGVNGAALDD746 pKa = 3.94NVNEE750 pKa = 4.4INWILNQDD758 pKa = 3.9FEE760 pKa = 5.59SDD762 pKa = 3.41GTGSVDD768 pKa = 3.33GKK770 pKa = 10.28FSGWEE775 pKa = 3.52VQYY778 pKa = 10.94AIWEE782 pKa = 4.26LTNNFPSDD790 pKa = 3.29AALNATPSNGQVEE803 pKa = 4.61DD804 pKa = 2.8VDD806 pKa = 4.88YY807 pKa = 10.75IVSRR811 pKa = 11.84AISEE815 pKa = 4.66GYY817 pKa = 10.39DD818 pKa = 3.35FEE820 pKa = 7.23AGVGDD825 pKa = 4.54IIGLVITPDD834 pKa = 3.23PVTAQNAQPFLVGVEE849 pKa = 4.09FEE851 pKa = 5.22TYY853 pKa = 10.71DD854 pKa = 3.8CLCC857 pKa = 4.56
Molecular weight: 92.98 kDa
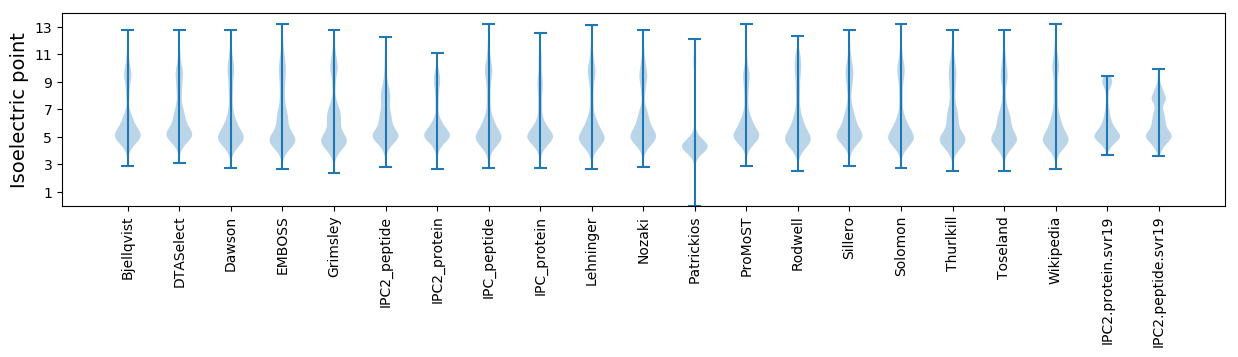
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3TXB8|A3TXB8_PSEBH Uncharacterized protein OS=Pseudooceanicola batsensis (strain ATCC BAA-863 / DSM 15984 / KCTC 12145 / HTCC2597) OX=252305 GN=OB2597_02622 PE=4 SV=1
MM1 pKa = 7.61ILRR4 pKa = 11.84RR5 pKa = 11.84VLSRR9 pKa = 11.84GLKK12 pKa = 10.02AGMGRR17 pKa = 11.84MAKK20 pKa = 10.32GDD22 pKa = 3.65GQGPQSKK29 pKa = 9.75RR30 pKa = 11.84ARR32 pKa = 11.84QTARR36 pKa = 11.84RR37 pKa = 11.84ARR39 pKa = 11.84KK40 pKa = 8.67AARR43 pKa = 11.84LMRR46 pKa = 11.84RR47 pKa = 11.84NGRR50 pKa = 11.84LL51 pKa = 3.09
MM1 pKa = 7.61ILRR4 pKa = 11.84RR5 pKa = 11.84VLSRR9 pKa = 11.84GLKK12 pKa = 10.02AGMGRR17 pKa = 11.84MAKK20 pKa = 10.32GDD22 pKa = 3.65GQGPQSKK29 pKa = 9.75RR30 pKa = 11.84ARR32 pKa = 11.84QTARR36 pKa = 11.84RR37 pKa = 11.84ARR39 pKa = 11.84KK40 pKa = 8.67AARR43 pKa = 11.84LMRR46 pKa = 11.84RR47 pKa = 11.84NGRR50 pKa = 11.84LL51 pKa = 3.09
Molecular weight: 5.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1317978 |
19 |
12228 |
314.3 |
34.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.325 ± 0.062 | 0.836 ± 0.018 |
6.337 ± 0.07 | 6.265 ± 0.033 |
3.618 ± 0.028 | 9.036 ± 0.055 |
2.025 ± 0.026 | 5.02 ± 0.028 |
2.706 ± 0.038 | 9.891 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.712 ± 0.027 | 2.407 ± 0.025 |
5.175 ± 0.034 | 2.953 ± 0.02 |
7.332 ± 0.058 | 5.0 ± 0.028 |
5.521 ± 0.045 | 7.334 ± 0.03 |
1.319 ± 0.016 | 2.189 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
