
Marinitenerispora sediminis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Marinitenerispora
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
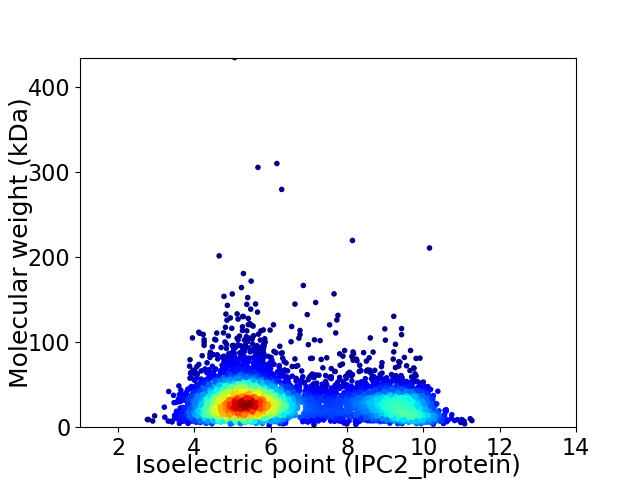
Virtual 2D-PAGE plot for 5230 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A368T6X3|A0A368T6X3_9ACTN AsnC family transcriptional regulator OS=Marinitenerispora sediminis OX=1931232 GN=DEF24_09175 PE=4 SV=1
MM1 pKa = 7.85RR2 pKa = 11.84SHH4 pKa = 6.81PVGRR8 pKa = 11.84VGAAAAAGTCLALLLTACGSGYY30 pKa = 10.99GDD32 pKa = 3.49TPEE35 pKa = 5.61GDD37 pKa = 3.47GALTVLFGSSGEE49 pKa = 4.22AEE51 pKa = 4.09TAAVKK56 pKa = 10.25AALAAWSEE64 pKa = 4.26EE65 pKa = 4.04SGVPAEE71 pKa = 5.3AIPAQDD77 pKa = 4.03LVQQLRR83 pKa = 11.84QGFAGGDD90 pKa = 3.67PADD93 pKa = 3.75VFYY96 pKa = 11.32VSPDD100 pKa = 3.16LFQQYY105 pKa = 11.21AEE107 pKa = 4.72GGSLYY112 pKa = 10.06PYY114 pKa = 10.09GDD116 pKa = 3.65QIEE119 pKa = 4.91DD120 pKa = 3.23VDD122 pKa = 4.36DD123 pKa = 4.97FYY125 pKa = 11.2PALRR129 pKa = 11.84DD130 pKa = 3.32SYY132 pKa = 10.66TYY134 pKa = 10.79EE135 pKa = 4.68DD136 pKa = 4.1EE137 pKa = 4.94LYY139 pKa = 10.67CVPKK143 pKa = 10.46DD144 pKa = 4.07YY145 pKa = 9.12NTHH148 pKa = 6.22ALVINTDD155 pKa = 2.96LWEE158 pKa = 4.2EE159 pKa = 4.21AGLGDD164 pKa = 5.71DD165 pKa = 6.16DD166 pKa = 6.2IPTSWEE172 pKa = 3.81EE173 pKa = 3.71LAEE176 pKa = 4.06TAEE179 pKa = 4.06QLTEE183 pKa = 3.86GDD185 pKa = 3.2RR186 pKa = 11.84VGFALNGDD194 pKa = 4.08YY195 pKa = 11.29NSAGTFMLQAGGWFVNDD212 pKa = 4.73DD213 pKa = 3.61NTEE216 pKa = 4.11ATGDD220 pKa = 3.74TPEE223 pKa = 4.38NLAALDD229 pKa = 3.96YY230 pKa = 9.56LTTGLDD236 pKa = 2.74EE237 pKa = 5.53GYY239 pKa = 10.7FSFVKK244 pKa = 10.73DD245 pKa = 2.97IDD247 pKa = 3.9AQSGSEE253 pKa = 3.71ALGRR257 pKa = 11.84GRR259 pKa = 11.84AAMVMDD265 pKa = 4.75GGWVTGAFDD274 pKa = 4.18NDD276 pKa = 3.63YY277 pKa = 11.47PEE279 pKa = 4.89LNWRR283 pKa = 11.84AVEE286 pKa = 4.02LPEE289 pKa = 5.22GPGGQATTVFSNCWGIAEE307 pKa = 4.4ASGDD311 pKa = 3.59HH312 pKa = 6.28EE313 pKa = 4.35AAVDD317 pKa = 3.74LVRR320 pKa = 11.84YY321 pKa = 7.33LTSPEE326 pKa = 3.82QQQAFAEE333 pKa = 4.47DD334 pKa = 4.72FGAVPPRR341 pKa = 11.84EE342 pKa = 4.15SLADD346 pKa = 3.29WTAEE350 pKa = 4.05TFPEE354 pKa = 4.29KK355 pKa = 10.54AAFAAGVDD363 pKa = 3.92YY364 pKa = 11.26ARR366 pKa = 11.84GQVPVPGFMSVLNEE380 pKa = 4.15FTTGMQGVAAGSAEE394 pKa = 4.07PADD397 pKa = 4.13VLAQLQRR404 pKa = 11.84DD405 pKa = 4.01GEE407 pKa = 4.35EE408 pKa = 4.24ALGEE412 pKa = 4.12
MM1 pKa = 7.85RR2 pKa = 11.84SHH4 pKa = 6.81PVGRR8 pKa = 11.84VGAAAAAGTCLALLLTACGSGYY30 pKa = 10.99GDD32 pKa = 3.49TPEE35 pKa = 5.61GDD37 pKa = 3.47GALTVLFGSSGEE49 pKa = 4.22AEE51 pKa = 4.09TAAVKK56 pKa = 10.25AALAAWSEE64 pKa = 4.26EE65 pKa = 4.04SGVPAEE71 pKa = 5.3AIPAQDD77 pKa = 4.03LVQQLRR83 pKa = 11.84QGFAGGDD90 pKa = 3.67PADD93 pKa = 3.75VFYY96 pKa = 11.32VSPDD100 pKa = 3.16LFQQYY105 pKa = 11.21AEE107 pKa = 4.72GGSLYY112 pKa = 10.06PYY114 pKa = 10.09GDD116 pKa = 3.65QIEE119 pKa = 4.91DD120 pKa = 3.23VDD122 pKa = 4.36DD123 pKa = 4.97FYY125 pKa = 11.2PALRR129 pKa = 11.84DD130 pKa = 3.32SYY132 pKa = 10.66TYY134 pKa = 10.79EE135 pKa = 4.68DD136 pKa = 4.1EE137 pKa = 4.94LYY139 pKa = 10.67CVPKK143 pKa = 10.46DD144 pKa = 4.07YY145 pKa = 9.12NTHH148 pKa = 6.22ALVINTDD155 pKa = 2.96LWEE158 pKa = 4.2EE159 pKa = 4.21AGLGDD164 pKa = 5.71DD165 pKa = 6.16DD166 pKa = 6.2IPTSWEE172 pKa = 3.81EE173 pKa = 3.71LAEE176 pKa = 4.06TAEE179 pKa = 4.06QLTEE183 pKa = 3.86GDD185 pKa = 3.2RR186 pKa = 11.84VGFALNGDD194 pKa = 4.08YY195 pKa = 11.29NSAGTFMLQAGGWFVNDD212 pKa = 4.73DD213 pKa = 3.61NTEE216 pKa = 4.11ATGDD220 pKa = 3.74TPEE223 pKa = 4.38NLAALDD229 pKa = 3.96YY230 pKa = 9.56LTTGLDD236 pKa = 2.74EE237 pKa = 5.53GYY239 pKa = 10.7FSFVKK244 pKa = 10.73DD245 pKa = 2.97IDD247 pKa = 3.9AQSGSEE253 pKa = 3.71ALGRR257 pKa = 11.84GRR259 pKa = 11.84AAMVMDD265 pKa = 4.75GGWVTGAFDD274 pKa = 4.18NDD276 pKa = 3.63YY277 pKa = 11.47PEE279 pKa = 4.89LNWRR283 pKa = 11.84AVEE286 pKa = 4.02LPEE289 pKa = 5.22GPGGQATTVFSNCWGIAEE307 pKa = 4.4ASGDD311 pKa = 3.59HH312 pKa = 6.28EE313 pKa = 4.35AAVDD317 pKa = 3.74LVRR320 pKa = 11.84YY321 pKa = 7.33LTSPEE326 pKa = 3.82QQQAFAEE333 pKa = 4.47DD334 pKa = 4.72FGAVPPRR341 pKa = 11.84EE342 pKa = 4.15SLADD346 pKa = 3.29WTAEE350 pKa = 4.05TFPEE354 pKa = 4.29KK355 pKa = 10.54AAFAAGVDD363 pKa = 3.92YY364 pKa = 11.26ARR366 pKa = 11.84GQVPVPGFMSVLNEE380 pKa = 4.15FTTGMQGVAAGSAEE394 pKa = 4.07PADD397 pKa = 4.13VLAQLQRR404 pKa = 11.84DD405 pKa = 4.01GEE407 pKa = 4.35EE408 pKa = 4.24ALGEE412 pKa = 4.12
Molecular weight: 43.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A368T1V2|A0A368T1V2_9ACTN Aldo/keto reductase OS=Marinitenerispora sediminis OX=1931232 GN=DEF24_19590 PE=4 SV=1
MM1 pKa = 7.78RR2 pKa = 11.84ALIAPPASASALSARR17 pKa = 11.84AARR20 pKa = 11.84VATLLRR26 pKa = 11.84VSATRR31 pKa = 11.84TSLTARR37 pKa = 11.84ARR39 pKa = 11.84SRR41 pKa = 11.84STSSRR46 pKa = 11.84LGVPPATPARR56 pKa = 11.84LSAAPALAALIRR68 pKa = 11.84VLKK71 pKa = 10.53RR72 pKa = 11.84WTTT75 pKa = 3.31
MM1 pKa = 7.78RR2 pKa = 11.84ALIAPPASASALSARR17 pKa = 11.84AARR20 pKa = 11.84VATLLRR26 pKa = 11.84VSATRR31 pKa = 11.84TSLTARR37 pKa = 11.84ARR39 pKa = 11.84SRR41 pKa = 11.84STSSRR46 pKa = 11.84LGVPPATPARR56 pKa = 11.84LSAAPALAALIRR68 pKa = 11.84VLKK71 pKa = 10.53RR72 pKa = 11.84WTTT75 pKa = 3.31
Molecular weight: 7.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1586353 |
22 |
3991 |
303.3 |
32.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.433 ± 0.057 | 0.748 ± 0.009 |
6.043 ± 0.034 | 5.977 ± 0.036 |
2.649 ± 0.021 | 9.597 ± 0.033 |
2.223 ± 0.017 | 3.081 ± 0.027 |
1.31 ± 0.023 | 10.354 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.693 ± 0.014 | 1.572 ± 0.015 |
6.347 ± 0.035 | 2.415 ± 0.022 |
9.124 ± 0.044 | 4.684 ± 0.024 |
5.488 ± 0.025 | 8.828 ± 0.032 |
1.494 ± 0.015 | 1.939 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
