
Plasmodium cynomolgi (strain B)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Plasmodium); Plasmodium cynomolgi
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
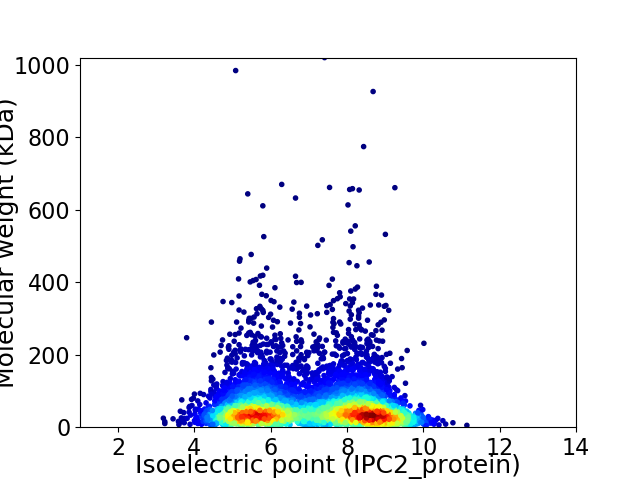
Virtual 2D-PAGE plot for 5713 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K6UJA2|K6UJA2_PLACD HIT-type domain-containing protein OS=Plasmodium cynomolgi (strain B) OX=1120755 GN=PCYB_072450 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.55NSLCLIFCLFFLNEE16 pKa = 4.18FACGEE21 pKa = 4.06RR22 pKa = 11.84EE23 pKa = 4.01EE24 pKa = 5.11TIDD27 pKa = 3.72AVPNVAVVNLEE38 pKa = 3.89KK39 pKa = 9.69PTLRR43 pKa = 11.84TLLMEE48 pKa = 4.4NPGVVGEE55 pKa = 4.25EE56 pKa = 4.11VSEE59 pKa = 4.52GVANGQVVNEE69 pKa = 4.55GVPSDD74 pKa = 4.64DD75 pKa = 4.49LPVDD79 pKa = 4.2RR80 pKa = 11.84VATEE84 pKa = 3.95EE85 pKa = 4.09VSPEE89 pKa = 3.87VAPSDD94 pKa = 4.12DD95 pKa = 3.98APSALSVEE103 pKa = 4.46TAEE106 pKa = 5.13GEE108 pKa = 4.3NQEE111 pKa = 4.99SEE113 pKa = 4.5QKK115 pKa = 9.51TPPDD119 pKa = 3.3NAKK122 pKa = 10.34QSDD125 pKa = 3.96ATPVEE130 pKa = 4.47VTHH133 pKa = 6.33GVNSVVEE140 pKa = 4.15NEE142 pKa = 4.22EE143 pKa = 4.12GKK145 pKa = 9.97HH146 pKa = 5.15VEE148 pKa = 3.9NDD150 pKa = 3.55SPVVTKK156 pKa = 10.4GQQEE160 pKa = 4.18EE161 pKa = 4.92GEE163 pKa = 4.46EE164 pKa = 4.35CCNEE168 pKa = 3.92CSTPCVPEE176 pKa = 4.06CTEE179 pKa = 4.29GDD181 pKa = 3.67NNCPEE186 pKa = 4.22KK187 pKa = 10.94CSTDD191 pKa = 3.58DD192 pKa = 4.62ACTEE196 pKa = 4.04DD197 pKa = 4.63CVVHH201 pKa = 6.02EE202 pKa = 4.4NCEE205 pKa = 4.09VADD208 pKa = 4.08CPEE211 pKa = 4.39CSGCPKK217 pKa = 10.65DD218 pKa = 4.24SACSEE223 pKa = 4.49CPTDD227 pKa = 5.19SEE229 pKa = 5.37CPQCSKK235 pKa = 10.84CPQCAEE241 pKa = 4.38CPQCSKK247 pKa = 10.93CPQCAEE253 pKa = 3.97NPQCSKK259 pKa = 10.85CPQCAEE265 pKa = 4.11CPEE268 pKa = 4.39CSKK271 pKa = 11.12CPEE274 pKa = 4.07CSKK277 pKa = 10.75CLEE280 pKa = 4.87DD281 pKa = 4.2STCDD285 pKa = 3.27EE286 pKa = 4.58CAKK289 pKa = 10.63CPEE292 pKa = 4.46DD293 pKa = 4.74PEE295 pKa = 4.37CAEE298 pKa = 4.64CKK300 pKa = 10.07RR301 pKa = 11.84EE302 pKa = 4.0CEE304 pKa = 3.95MRR306 pKa = 11.84TEE308 pKa = 3.97EE309 pKa = 5.32GEE311 pKa = 4.1NQVAEE316 pKa = 4.32EE317 pKa = 4.32RR318 pKa = 11.84TSEE321 pKa = 4.61DD322 pKa = 2.93SPQTVDD328 pKa = 3.26TPHH331 pKa = 7.52VEE333 pKa = 4.38VVNSVDD339 pKa = 3.53PVAEE343 pKa = 4.04EE344 pKa = 4.73AEE346 pKa = 4.56GKK348 pKa = 9.89QPEE351 pKa = 4.45IEE353 pKa = 4.56DD354 pKa = 3.65NSDD357 pKa = 3.41GEE359 pKa = 4.42GANNVIRR366 pKa = 11.84EE367 pKa = 4.19IFNGLKK373 pKa = 10.22NVEE376 pKa = 4.37DD377 pKa = 4.0EE378 pKa = 4.53EE379 pKa = 4.51EE380 pKa = 4.41TVLEE384 pKa = 4.23EE385 pKa = 4.03TDD387 pKa = 3.78EE388 pKa = 4.45EE389 pKa = 4.67EE390 pKa = 4.48GDD392 pKa = 3.74QMVDD396 pKa = 3.11EE397 pKa = 4.76NEE399 pKa = 4.06EE400 pKa = 4.07EE401 pKa = 4.55EE402 pKa = 4.35EE403 pKa = 4.66GEE405 pKa = 4.38TYY407 pKa = 10.45EE408 pKa = 5.83GEE410 pKa = 4.22DD411 pKa = 3.91TEE413 pKa = 6.03DD414 pKa = 3.36VALDD418 pKa = 3.71EE419 pKa = 4.62EE420 pKa = 4.79DD421 pKa = 3.93EE422 pKa = 4.31EE423 pKa = 5.13GEE425 pKa = 3.95KK426 pKa = 11.03DD427 pKa = 3.64EE428 pKa = 4.51EE429 pKa = 4.54GKK431 pKa = 10.72AGEE434 pKa = 4.44EE435 pKa = 4.45GNAGEE440 pKa = 4.34GGKK443 pKa = 10.39AGEE446 pKa = 4.08EE447 pKa = 4.29GKK449 pKa = 10.64AGEE452 pKa = 4.15EE453 pKa = 4.37GKK455 pKa = 10.58AGEE458 pKa = 4.4QSVEE462 pKa = 4.04EE463 pKa = 4.2QNAEE467 pKa = 3.96VPFIDD472 pKa = 3.86TATNNKK478 pKa = 8.54GVRR481 pKa = 11.84NIKK484 pKa = 9.36TEE486 pKa = 3.92THH488 pKa = 6.23NLVADD493 pKa = 5.81LITILNADD501 pKa = 3.62NGIDD505 pKa = 3.61QSLKK509 pKa = 11.0ALAKK513 pKa = 10.85DD514 pKa = 3.2MAQYY518 pKa = 10.62FLNHH522 pKa = 6.31LNAEE526 pKa = 4.14NEE528 pKa = 4.15IAA530 pKa = 4.82
MM1 pKa = 7.54KK2 pKa = 10.55NSLCLIFCLFFLNEE16 pKa = 4.18FACGEE21 pKa = 4.06RR22 pKa = 11.84EE23 pKa = 4.01EE24 pKa = 5.11TIDD27 pKa = 3.72AVPNVAVVNLEE38 pKa = 3.89KK39 pKa = 9.69PTLRR43 pKa = 11.84TLLMEE48 pKa = 4.4NPGVVGEE55 pKa = 4.25EE56 pKa = 4.11VSEE59 pKa = 4.52GVANGQVVNEE69 pKa = 4.55GVPSDD74 pKa = 4.64DD75 pKa = 4.49LPVDD79 pKa = 4.2RR80 pKa = 11.84VATEE84 pKa = 3.95EE85 pKa = 4.09VSPEE89 pKa = 3.87VAPSDD94 pKa = 4.12DD95 pKa = 3.98APSALSVEE103 pKa = 4.46TAEE106 pKa = 5.13GEE108 pKa = 4.3NQEE111 pKa = 4.99SEE113 pKa = 4.5QKK115 pKa = 9.51TPPDD119 pKa = 3.3NAKK122 pKa = 10.34QSDD125 pKa = 3.96ATPVEE130 pKa = 4.47VTHH133 pKa = 6.33GVNSVVEE140 pKa = 4.15NEE142 pKa = 4.22EE143 pKa = 4.12GKK145 pKa = 9.97HH146 pKa = 5.15VEE148 pKa = 3.9NDD150 pKa = 3.55SPVVTKK156 pKa = 10.4GQQEE160 pKa = 4.18EE161 pKa = 4.92GEE163 pKa = 4.46EE164 pKa = 4.35CCNEE168 pKa = 3.92CSTPCVPEE176 pKa = 4.06CTEE179 pKa = 4.29GDD181 pKa = 3.67NNCPEE186 pKa = 4.22KK187 pKa = 10.94CSTDD191 pKa = 3.58DD192 pKa = 4.62ACTEE196 pKa = 4.04DD197 pKa = 4.63CVVHH201 pKa = 6.02EE202 pKa = 4.4NCEE205 pKa = 4.09VADD208 pKa = 4.08CPEE211 pKa = 4.39CSGCPKK217 pKa = 10.65DD218 pKa = 4.24SACSEE223 pKa = 4.49CPTDD227 pKa = 5.19SEE229 pKa = 5.37CPQCSKK235 pKa = 10.84CPQCAEE241 pKa = 4.38CPQCSKK247 pKa = 10.93CPQCAEE253 pKa = 3.97NPQCSKK259 pKa = 10.85CPQCAEE265 pKa = 4.11CPEE268 pKa = 4.39CSKK271 pKa = 11.12CPEE274 pKa = 4.07CSKK277 pKa = 10.75CLEE280 pKa = 4.87DD281 pKa = 4.2STCDD285 pKa = 3.27EE286 pKa = 4.58CAKK289 pKa = 10.63CPEE292 pKa = 4.46DD293 pKa = 4.74PEE295 pKa = 4.37CAEE298 pKa = 4.64CKK300 pKa = 10.07RR301 pKa = 11.84EE302 pKa = 4.0CEE304 pKa = 3.95MRR306 pKa = 11.84TEE308 pKa = 3.97EE309 pKa = 5.32GEE311 pKa = 4.1NQVAEE316 pKa = 4.32EE317 pKa = 4.32RR318 pKa = 11.84TSEE321 pKa = 4.61DD322 pKa = 2.93SPQTVDD328 pKa = 3.26TPHH331 pKa = 7.52VEE333 pKa = 4.38VVNSVDD339 pKa = 3.53PVAEE343 pKa = 4.04EE344 pKa = 4.73AEE346 pKa = 4.56GKK348 pKa = 9.89QPEE351 pKa = 4.45IEE353 pKa = 4.56DD354 pKa = 3.65NSDD357 pKa = 3.41GEE359 pKa = 4.42GANNVIRR366 pKa = 11.84EE367 pKa = 4.19IFNGLKK373 pKa = 10.22NVEE376 pKa = 4.37DD377 pKa = 4.0EE378 pKa = 4.53EE379 pKa = 4.51EE380 pKa = 4.41TVLEE384 pKa = 4.23EE385 pKa = 4.03TDD387 pKa = 3.78EE388 pKa = 4.45EE389 pKa = 4.67EE390 pKa = 4.48GDD392 pKa = 3.74QMVDD396 pKa = 3.11EE397 pKa = 4.76NEE399 pKa = 4.06EE400 pKa = 4.07EE401 pKa = 4.55EE402 pKa = 4.35EE403 pKa = 4.66GEE405 pKa = 4.38TYY407 pKa = 10.45EE408 pKa = 5.83GEE410 pKa = 4.22DD411 pKa = 3.91TEE413 pKa = 6.03DD414 pKa = 3.36VALDD418 pKa = 3.71EE419 pKa = 4.62EE420 pKa = 4.79DD421 pKa = 3.93EE422 pKa = 4.31EE423 pKa = 5.13GEE425 pKa = 3.95KK426 pKa = 11.03DD427 pKa = 3.64EE428 pKa = 4.51EE429 pKa = 4.54GKK431 pKa = 10.72AGEE434 pKa = 4.44EE435 pKa = 4.45GNAGEE440 pKa = 4.34GGKK443 pKa = 10.39AGEE446 pKa = 4.08EE447 pKa = 4.29GKK449 pKa = 10.64AGEE452 pKa = 4.15EE453 pKa = 4.37GKK455 pKa = 10.58AGEE458 pKa = 4.4QSVEE462 pKa = 4.04EE463 pKa = 4.2QNAEE467 pKa = 3.96VPFIDD472 pKa = 3.86TATNNKK478 pKa = 8.54GVRR481 pKa = 11.84NIKK484 pKa = 9.36TEE486 pKa = 3.92THH488 pKa = 6.23NLVADD493 pKa = 5.81LITILNADD501 pKa = 3.62NGIDD505 pKa = 3.61QSLKK509 pKa = 11.0ALAKK513 pKa = 10.85DD514 pKa = 3.2MAQYY518 pKa = 10.62FLNHH522 pKa = 6.31LNAEE526 pKa = 4.14NEE528 pKa = 4.15IAA530 pKa = 4.82
Molecular weight: 57.15 kDa
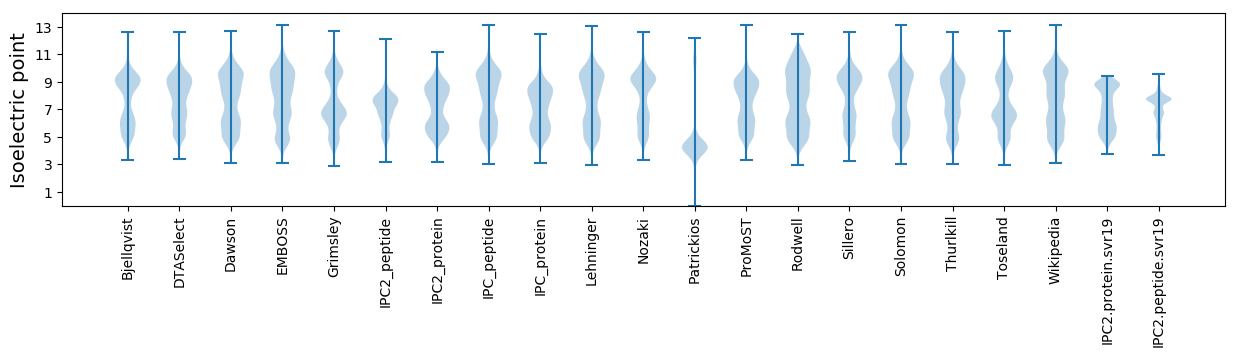
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K6UWW3|K6UWW3_PLACD DNA helicase OS=Plasmodium cynomolgi (strain B) OX=1120755 GN=PCYB_102750 PE=4 SV=1
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3284714 |
16 |
8883 |
575.0 |
65.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.437 ± 0.029 | 1.892 ± 0.016 |
5.74 ± 0.026 | 7.705 ± 0.048 |
4.28 ± 0.03 | 5.966 ± 0.051 |
2.639 ± 0.016 | 6.229 ± 0.04 |
9.364 ± 0.039 | 8.129 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.158 ± 0.011 | 8.02 ± 0.046 |
3.117 ± 0.025 | 3.198 ± 0.016 |
4.561 ± 0.034 | 7.982 ± 0.035 |
4.497 ± 0.018 | 5.161 ± 0.02 |
0.632 ± 0.008 | 4.287 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
