
Halovirus HSTV-1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified archaeal dsDNA viruses; Haloviruses
Average proteome isoelectric point is 4.81
Get precalculated fractions of proteins
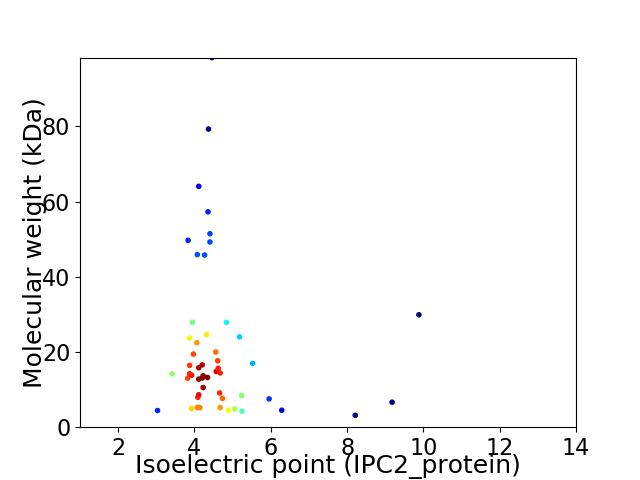
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9QTN4|R9QTN4_9VIRU PCNA protein OS=Halovirus HSTV-1 OX=1262530 GN=40 PE=3 SV=1
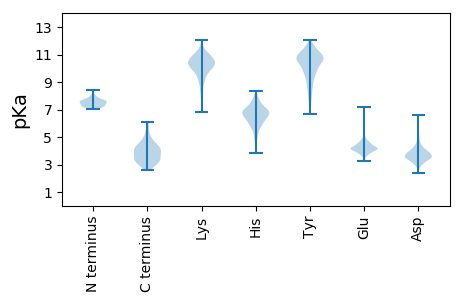
MM1 pKa = 7.64TDD3 pKa = 3.15GAVDD7 pKa = 3.48ADD9 pKa = 4.13YY10 pKa = 11.78GEE12 pKa = 4.77GLQDD16 pKa = 3.45YY17 pKa = 9.48YY18 pKa = 11.8DD19 pKa = 3.88RR20 pKa = 11.84VADD23 pKa = 4.03ALEE26 pKa = 4.55DD27 pKa = 3.84VPGEE31 pKa = 4.05PVVGGCAIDD40 pKa = 3.11IVTRR44 pKa = 11.84QVVYY48 pKa = 10.86VVDD51 pKa = 5.05RR52 pKa = 11.84VADD55 pKa = 3.83SCVEE59 pKa = 4.04YY60 pKa = 11.23YY61 pKa = 10.58DD62 pKa = 5.47AEE64 pKa = 4.47GFDD67 pKa = 3.56LVTYY71 pKa = 10.44KK72 pKa = 9.01MHH74 pKa = 7.22PFLPGISVEE83 pKa = 4.19NAVYY87 pKa = 10.27EE88 pKa = 4.34CVYY91 pKa = 11.15VDD93 pKa = 4.42GNPQNTHH100 pKa = 5.66KK101 pKa = 10.68PGRR104 pKa = 11.84TYY106 pKa = 11.11DD107 pKa = 3.5FPTARR112 pKa = 11.84LMPFPVGIATDD123 pKa = 3.71TYY125 pKa = 11.04EE126 pKa = 4.08VGGVV130 pKa = 3.17
MM1 pKa = 7.64TDD3 pKa = 3.15GAVDD7 pKa = 3.48ADD9 pKa = 4.13YY10 pKa = 11.78GEE12 pKa = 4.77GLQDD16 pKa = 3.45YY17 pKa = 9.48YY18 pKa = 11.8DD19 pKa = 3.88RR20 pKa = 11.84VADD23 pKa = 4.03ALEE26 pKa = 4.55DD27 pKa = 3.84VPGEE31 pKa = 4.05PVVGGCAIDD40 pKa = 3.11IVTRR44 pKa = 11.84QVVYY48 pKa = 10.86VVDD51 pKa = 5.05RR52 pKa = 11.84VADD55 pKa = 3.83SCVEE59 pKa = 4.04YY60 pKa = 11.23YY61 pKa = 10.58DD62 pKa = 5.47AEE64 pKa = 4.47GFDD67 pKa = 3.56LVTYY71 pKa = 10.44KK72 pKa = 9.01MHH74 pKa = 7.22PFLPGISVEE83 pKa = 4.19NAVYY87 pKa = 10.27EE88 pKa = 4.34CVYY91 pKa = 11.15VDD93 pKa = 4.42GNPQNTHH100 pKa = 5.66KK101 pKa = 10.68PGRR104 pKa = 11.84TYY106 pKa = 11.11DD107 pKa = 3.5FPTARR112 pKa = 11.84LMPFPVGIATDD123 pKa = 3.71TYY125 pKa = 11.04EE126 pKa = 4.08VGGVV130 pKa = 3.17
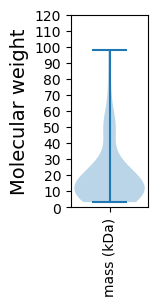
Molecular weight: 14.21 kDa
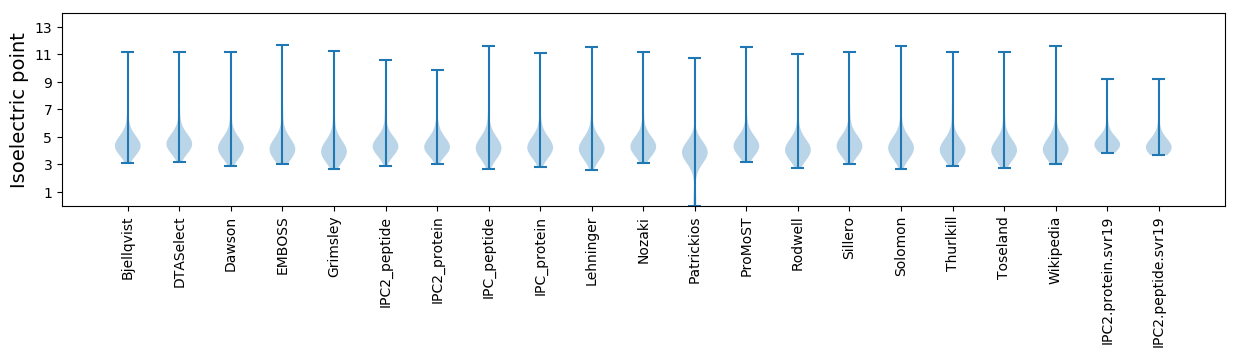
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9QTM6|R9QTM6_9VIRU Uncharacterized protein OS=Halovirus HSTV-1 OX=1262530 GN=5 PE=4 SV=1
MM1 pKa = 7.53AVGRR5 pKa = 11.84SHH7 pKa = 6.85TPHH10 pKa = 6.5TAGSAVDD17 pKa = 3.45AAGDD21 pKa = 3.43VSGNVSRR28 pKa = 11.84TAPRR32 pKa = 11.84EE33 pKa = 4.01DD34 pKa = 3.63RR35 pKa = 11.84RR36 pKa = 11.84GTAPNRR42 pKa = 11.84QTYY45 pKa = 9.97AIWPTMKK52 pKa = 9.41TCRR55 pKa = 11.84ATWTRR60 pKa = 11.84HH61 pKa = 4.67SLSPTIGLRR70 pKa = 11.84SHH72 pKa = 6.49HH73 pKa = 5.86TTPILEE79 pKa = 4.3RR80 pKa = 11.84VGHH83 pKa = 5.98SSGFDD88 pKa = 3.21SLSLRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84LRR97 pKa = 11.84KK98 pKa = 9.79GVWIRR103 pKa = 11.84SWARR107 pKa = 11.84FFRR110 pKa = 11.84VVSPRR115 pKa = 11.84PYY117 pKa = 8.06NTRR120 pKa = 11.84VKK122 pKa = 9.8TRR124 pKa = 11.84AEE126 pKa = 3.84RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84HH130 pKa = 4.89FSHH133 pKa = 6.98TFRR136 pKa = 11.84LAYY139 pKa = 9.09MNHH142 pKa = 7.15PALCSISHH150 pKa = 6.85DD151 pKa = 3.64CVVDD155 pKa = 4.24VMATGQSRR163 pKa = 11.84TTDD166 pKa = 3.28KK167 pKa = 10.98PSSTDD172 pKa = 3.13YY173 pKa = 10.73RR174 pKa = 11.84NCLITPMSSDD184 pKa = 3.48PPLIRR189 pKa = 11.84PEE191 pKa = 4.07HH192 pKa = 6.6VIDD195 pKa = 4.16ALLVGLLAFFGVILADD211 pKa = 3.48VLTALLQGRR220 pKa = 11.84VVYY223 pKa = 8.47LTVSDD228 pKa = 3.73IAARR232 pKa = 11.84TPTGVVAFGLTFVFQWARR250 pKa = 11.84ARR252 pKa = 11.84GIDD255 pKa = 3.39VLAAYY260 pKa = 9.96RR261 pKa = 11.84KK262 pKa = 10.03FKK264 pKa = 10.94DD265 pKa = 3.4SLPP268 pKa = 3.84
MM1 pKa = 7.53AVGRR5 pKa = 11.84SHH7 pKa = 6.85TPHH10 pKa = 6.5TAGSAVDD17 pKa = 3.45AAGDD21 pKa = 3.43VSGNVSRR28 pKa = 11.84TAPRR32 pKa = 11.84EE33 pKa = 4.01DD34 pKa = 3.63RR35 pKa = 11.84RR36 pKa = 11.84GTAPNRR42 pKa = 11.84QTYY45 pKa = 9.97AIWPTMKK52 pKa = 9.41TCRR55 pKa = 11.84ATWTRR60 pKa = 11.84HH61 pKa = 4.67SLSPTIGLRR70 pKa = 11.84SHH72 pKa = 6.49HH73 pKa = 5.86TTPILEE79 pKa = 4.3RR80 pKa = 11.84VGHH83 pKa = 5.98SSGFDD88 pKa = 3.21SLSLRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84LRR97 pKa = 11.84KK98 pKa = 9.79GVWIRR103 pKa = 11.84SWARR107 pKa = 11.84FFRR110 pKa = 11.84VVSPRR115 pKa = 11.84PYY117 pKa = 8.06NTRR120 pKa = 11.84VKK122 pKa = 9.8TRR124 pKa = 11.84AEE126 pKa = 3.84RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84HH130 pKa = 4.89FSHH133 pKa = 6.98TFRR136 pKa = 11.84LAYY139 pKa = 9.09MNHH142 pKa = 7.15PALCSISHH150 pKa = 6.85DD151 pKa = 3.64CVVDD155 pKa = 4.24VMATGQSRR163 pKa = 11.84TTDD166 pKa = 3.28KK167 pKa = 10.98PSSTDD172 pKa = 3.13YY173 pKa = 10.73RR174 pKa = 11.84NCLITPMSSDD184 pKa = 3.48PPLIRR189 pKa = 11.84PEE191 pKa = 4.07HH192 pKa = 6.6VIDD195 pKa = 4.16ALLVGLLAFFGVILADD211 pKa = 3.48VLTALLQGRR220 pKa = 11.84VVYY223 pKa = 8.47LTVSDD228 pKa = 3.73IAARR232 pKa = 11.84TPTGVVAFGLTFVFQWARR250 pKa = 11.84ARR252 pKa = 11.84GIDD255 pKa = 3.39VLAAYY260 pKa = 9.96RR261 pKa = 11.84KK262 pKa = 10.03FKK264 pKa = 10.94DD265 pKa = 3.4SLPP268 pKa = 3.84
Molecular weight: 29.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10139 |
28 |
881 |
191.3 |
21.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.459 ± 0.322 | 0.937 ± 0.131 |
9.814 ± 0.285 | 8.334 ± 0.507 |
2.673 ± 0.181 | 7.792 ± 0.338 |
1.825 ± 0.18 | 4.458 ± 0.358 |
2.821 ± 0.198 | 6.687 ± 0.328 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.121 ± 0.15 | 3.639 ± 0.34 |
4.379 ± 0.209 | 3.61 ± 0.189 |
6.076 ± 0.295 | 6.283 ± 0.323 |
7.299 ± 0.396 | 7.427 ± 0.285 |
1.696 ± 0.14 | 2.673 ± 0.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
