
Colwellia sp. MT41
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Colwelliaceae; Colwellia; unclassified Colwellia
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
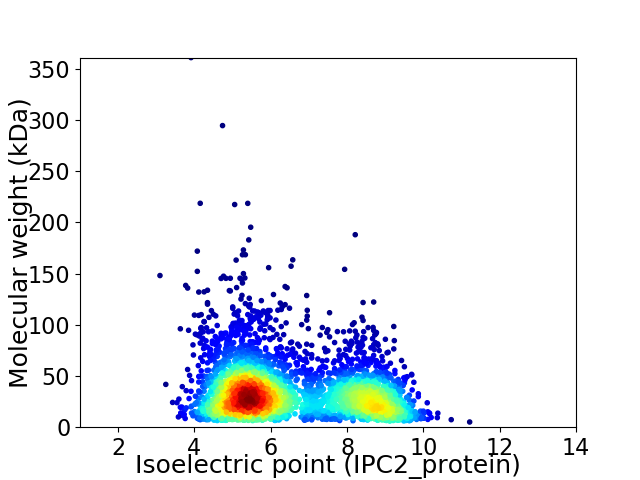
Virtual 2D-PAGE plot for 3491 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2JHN5|A0A0S2JHN5_9GAMM Uncharacterized protein OS=Colwellia sp. MT41 OX=58049 GN=CMT41_12295 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 10.55VIMDD6 pKa = 5.33KK7 pKa = 10.54ISKK10 pKa = 10.35CFGQYY15 pKa = 8.94RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 10.82ALFATMILVLSLSGCGGSSYY38 pKa = 11.46GDD40 pKa = 3.45DD41 pKa = 3.86KK42 pKa = 11.53VTVDD46 pKa = 3.79NVAPIITLIGDD57 pKa = 3.43AEE59 pKa = 4.28VRR61 pKa = 11.84VLVNGSYY68 pKa = 10.58TDD70 pKa = 3.89AGASANDD77 pKa = 3.58NVDD80 pKa = 3.05GKK82 pKa = 10.82ISVAITGSVDD92 pKa = 2.71TMTAGSYY99 pKa = 10.38ILTYY103 pKa = 9.89TATDD107 pKa = 3.55YY108 pKa = 11.54AGNVSTLTRR117 pKa = 11.84TVTVVPPTLTGTAAAGAAIVGTVTIKK143 pKa = 10.99DD144 pKa = 3.63SLGEE148 pKa = 4.08TRR150 pKa = 11.84SEE152 pKa = 4.34LIEE155 pKa = 5.34ADD157 pKa = 3.04GTYY160 pKa = 10.7SVDD163 pKa = 3.56VTGLTAPFRR172 pKa = 11.84LRR174 pKa = 11.84AEE176 pKa = 4.17GTVGGKK182 pKa = 8.41SYY184 pKa = 10.41RR185 pKa = 11.84IHH187 pKa = 7.39SYY189 pKa = 11.23AEE191 pKa = 3.76EE192 pKa = 4.05ATLDD196 pKa = 3.43GTINITPFTDD206 pKa = 5.38LIIANAAHH214 pKa = 6.34QIAAAYY220 pKa = 9.08FDD222 pKa = 4.3EE223 pKa = 4.58VSPTFLDD230 pKa = 3.68PVEE233 pKa = 4.47IAAQEE238 pKa = 4.18DD239 pKa = 3.85ALQAKK244 pKa = 8.72LQAVFDD250 pKa = 4.37ALGLDD255 pKa = 3.77SAINLLTSAFSADD268 pKa = 3.69HH269 pKa = 6.58SGLDD273 pKa = 3.34AALDD277 pKa = 4.25IIQIEE282 pKa = 4.34TDD284 pKa = 4.3PITNIATIVNVLDD297 pKa = 4.18GSSIEE302 pKa = 5.41DD303 pKa = 5.46DD304 pKa = 3.58ITDD307 pKa = 3.44SDD309 pKa = 4.42DD310 pKa = 3.73NEE312 pKa = 4.36SVIIVDD318 pKa = 3.99SAALTIVVTDD328 pKa = 3.91TQAIAALFASFAAEE342 pKa = 4.11FTDD345 pKa = 4.44GLPSLASLDD354 pKa = 3.86GFFADD359 pKa = 4.88SFLHH363 pKa = 6.47EE364 pKa = 5.14DD365 pKa = 3.3QAKK368 pKa = 9.95SQFLTDD374 pKa = 3.21ITTDD378 pKa = 3.31PSLIGLVFSSIAVRR392 pKa = 11.84DD393 pKa = 4.08LDD395 pKa = 4.55SMAGTATVDD404 pKa = 3.65FNVYY408 pKa = 9.96INGIVDD414 pKa = 3.69VEE416 pKa = 4.44VEE418 pKa = 3.9TWLLQKK424 pKa = 10.73DD425 pKa = 4.32SSQAWQLLGNQRR437 pKa = 11.84FYY439 pKa = 11.26EE440 pKa = 4.58ADD442 pKa = 3.54LLTFHH447 pKa = 7.18CNDD450 pKa = 3.62FDD452 pKa = 5.88GSDD455 pKa = 4.1DD456 pKa = 3.7MAGGCGLNVSFYY468 pKa = 11.56DD469 pKa = 4.3NDD471 pKa = 3.35FSNNGTNDD479 pKa = 3.08APIASGTMTIIDD491 pKa = 3.97GTDD494 pKa = 3.05GEE496 pKa = 4.84TVKK499 pKa = 11.02DD500 pKa = 3.41IVYY503 pKa = 10.23LGNPDD508 pKa = 3.83YY509 pKa = 10.89IAAGEE514 pKa = 4.08LQVYY518 pKa = 8.88NQTTQMYY525 pKa = 8.07QWDD528 pKa = 3.98YY529 pKa = 11.75AGFGMSDD536 pKa = 3.77GEE538 pKa = 4.08VDD540 pKa = 3.46PNIFVVGDD548 pKa = 3.53IIRR551 pKa = 11.84YY552 pKa = 9.0KK553 pKa = 10.83LYY555 pKa = 10.47TEE557 pKa = 4.12QLDD560 pKa = 3.93LTVPGQPMVSSGNEE574 pKa = 3.23VAMFDD579 pKa = 4.54KK580 pKa = 10.74ILMYY584 pKa = 10.75LPEE587 pKa = 4.15TTGRR591 pKa = 11.84YY592 pKa = 7.56PALTTDD598 pKa = 4.94GLTALNNFTLDD609 pKa = 3.17EE610 pKa = 4.93DD611 pKa = 4.48LTVSWTLQPGTVIDD625 pKa = 4.7SIWVEE630 pKa = 3.66ISDD633 pKa = 4.09NQGNYY638 pKa = 10.14YY639 pKa = 10.52DD640 pKa = 4.96VNDD643 pKa = 4.11KK644 pKa = 11.11SIAPDD649 pKa = 3.52VTSTTVDD656 pKa = 2.97SSVFSQDD663 pKa = 4.02LLNDD667 pKa = 3.81LDD669 pKa = 4.47FDD671 pKa = 4.12QTNMTLLVRR680 pKa = 11.84IYY682 pKa = 10.56SIVPTTGQTHH692 pKa = 4.76STDD695 pKa = 3.22YY696 pKa = 10.94RR697 pKa = 11.84RR698 pKa = 11.84TFAGTMPGDD707 pKa = 3.57GTQPGDD713 pKa = 3.23GTATVMCNTEE723 pKa = 4.53SPWDD727 pKa = 3.94DD728 pKa = 5.02ANDD731 pKa = 3.78KK732 pKa = 9.04PTVFYY737 pKa = 10.71SINDD741 pKa = 3.51FEE743 pKa = 4.62TAVADD748 pKa = 4.2CASQASLLNFTKK760 pKa = 10.84ANLAGLTWYY769 pKa = 10.89VEE771 pKa = 4.2DD772 pKa = 4.0EE773 pKa = 4.59RR774 pKa = 11.84IVFDD778 pKa = 3.79SSADD782 pKa = 3.49TFVITTYY789 pKa = 11.41GDD791 pKa = 3.62DD792 pKa = 4.0GEE794 pKa = 5.61LGGGDD799 pKa = 4.66DD800 pKa = 3.57EE801 pKa = 4.79TFYY804 pKa = 10.8GTVADD809 pKa = 4.01YY810 pKa = 7.92ATNVIEE816 pKa = 4.86FSFSFTPGGDD826 pKa = 2.63IVGRR830 pKa = 11.84DD831 pKa = 4.15LIRR834 pKa = 11.84VKK836 pKa = 10.89AEE838 pKa = 3.95SSFQGKK844 pKa = 10.08VIFTAVLLWEE854 pKa = 4.86FYY856 pKa = 11.02DD857 pKa = 3.57WAEE860 pKa = 4.17SDD862 pKa = 4.14GDD864 pKa = 4.33NIDD867 pKa = 3.9KK868 pKa = 10.79AHH870 pKa = 6.4QGGEE874 pKa = 4.06LRR876 pKa = 11.84TAGYY880 pKa = 10.56AVDD883 pKa = 5.9ADD885 pKa = 4.29FDD887 pKa = 3.91WAAWQATPP895 pKa = 4.9
MM1 pKa = 7.28KK2 pKa = 10.55VIMDD6 pKa = 5.33KK7 pKa = 10.54ISKK10 pKa = 10.35CFGQYY15 pKa = 8.94RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 10.82ALFATMILVLSLSGCGGSSYY38 pKa = 11.46GDD40 pKa = 3.45DD41 pKa = 3.86KK42 pKa = 11.53VTVDD46 pKa = 3.79NVAPIITLIGDD57 pKa = 3.43AEE59 pKa = 4.28VRR61 pKa = 11.84VLVNGSYY68 pKa = 10.58TDD70 pKa = 3.89AGASANDD77 pKa = 3.58NVDD80 pKa = 3.05GKK82 pKa = 10.82ISVAITGSVDD92 pKa = 2.71TMTAGSYY99 pKa = 10.38ILTYY103 pKa = 9.89TATDD107 pKa = 3.55YY108 pKa = 11.54AGNVSTLTRR117 pKa = 11.84TVTVVPPTLTGTAAAGAAIVGTVTIKK143 pKa = 10.99DD144 pKa = 3.63SLGEE148 pKa = 4.08TRR150 pKa = 11.84SEE152 pKa = 4.34LIEE155 pKa = 5.34ADD157 pKa = 3.04GTYY160 pKa = 10.7SVDD163 pKa = 3.56VTGLTAPFRR172 pKa = 11.84LRR174 pKa = 11.84AEE176 pKa = 4.17GTVGGKK182 pKa = 8.41SYY184 pKa = 10.41RR185 pKa = 11.84IHH187 pKa = 7.39SYY189 pKa = 11.23AEE191 pKa = 3.76EE192 pKa = 4.05ATLDD196 pKa = 3.43GTINITPFTDD206 pKa = 5.38LIIANAAHH214 pKa = 6.34QIAAAYY220 pKa = 9.08FDD222 pKa = 4.3EE223 pKa = 4.58VSPTFLDD230 pKa = 3.68PVEE233 pKa = 4.47IAAQEE238 pKa = 4.18DD239 pKa = 3.85ALQAKK244 pKa = 8.72LQAVFDD250 pKa = 4.37ALGLDD255 pKa = 3.77SAINLLTSAFSADD268 pKa = 3.69HH269 pKa = 6.58SGLDD273 pKa = 3.34AALDD277 pKa = 4.25IIQIEE282 pKa = 4.34TDD284 pKa = 4.3PITNIATIVNVLDD297 pKa = 4.18GSSIEE302 pKa = 5.41DD303 pKa = 5.46DD304 pKa = 3.58ITDD307 pKa = 3.44SDD309 pKa = 4.42DD310 pKa = 3.73NEE312 pKa = 4.36SVIIVDD318 pKa = 3.99SAALTIVVTDD328 pKa = 3.91TQAIAALFASFAAEE342 pKa = 4.11FTDD345 pKa = 4.44GLPSLASLDD354 pKa = 3.86GFFADD359 pKa = 4.88SFLHH363 pKa = 6.47EE364 pKa = 5.14DD365 pKa = 3.3QAKK368 pKa = 9.95SQFLTDD374 pKa = 3.21ITTDD378 pKa = 3.31PSLIGLVFSSIAVRR392 pKa = 11.84DD393 pKa = 4.08LDD395 pKa = 4.55SMAGTATVDD404 pKa = 3.65FNVYY408 pKa = 9.96INGIVDD414 pKa = 3.69VEE416 pKa = 4.44VEE418 pKa = 3.9TWLLQKK424 pKa = 10.73DD425 pKa = 4.32SSQAWQLLGNQRR437 pKa = 11.84FYY439 pKa = 11.26EE440 pKa = 4.58ADD442 pKa = 3.54LLTFHH447 pKa = 7.18CNDD450 pKa = 3.62FDD452 pKa = 5.88GSDD455 pKa = 4.1DD456 pKa = 3.7MAGGCGLNVSFYY468 pKa = 11.56DD469 pKa = 4.3NDD471 pKa = 3.35FSNNGTNDD479 pKa = 3.08APIASGTMTIIDD491 pKa = 3.97GTDD494 pKa = 3.05GEE496 pKa = 4.84TVKK499 pKa = 11.02DD500 pKa = 3.41IVYY503 pKa = 10.23LGNPDD508 pKa = 3.83YY509 pKa = 10.89IAAGEE514 pKa = 4.08LQVYY518 pKa = 8.88NQTTQMYY525 pKa = 8.07QWDD528 pKa = 3.98YY529 pKa = 11.75AGFGMSDD536 pKa = 3.77GEE538 pKa = 4.08VDD540 pKa = 3.46PNIFVVGDD548 pKa = 3.53IIRR551 pKa = 11.84YY552 pKa = 9.0KK553 pKa = 10.83LYY555 pKa = 10.47TEE557 pKa = 4.12QLDD560 pKa = 3.93LTVPGQPMVSSGNEE574 pKa = 3.23VAMFDD579 pKa = 4.54KK580 pKa = 10.74ILMYY584 pKa = 10.75LPEE587 pKa = 4.15TTGRR591 pKa = 11.84YY592 pKa = 7.56PALTTDD598 pKa = 4.94GLTALNNFTLDD609 pKa = 3.17EE610 pKa = 4.93DD611 pKa = 4.48LTVSWTLQPGTVIDD625 pKa = 4.7SIWVEE630 pKa = 3.66ISDD633 pKa = 4.09NQGNYY638 pKa = 10.14YY639 pKa = 10.52DD640 pKa = 4.96VNDD643 pKa = 4.11KK644 pKa = 11.11SIAPDD649 pKa = 3.52VTSTTVDD656 pKa = 2.97SSVFSQDD663 pKa = 4.02LLNDD667 pKa = 3.81LDD669 pKa = 4.47FDD671 pKa = 4.12QTNMTLLVRR680 pKa = 11.84IYY682 pKa = 10.56SIVPTTGQTHH692 pKa = 4.76STDD695 pKa = 3.22YY696 pKa = 10.94RR697 pKa = 11.84RR698 pKa = 11.84TFAGTMPGDD707 pKa = 3.57GTQPGDD713 pKa = 3.23GTATVMCNTEE723 pKa = 4.53SPWDD727 pKa = 3.94DD728 pKa = 5.02ANDD731 pKa = 3.78KK732 pKa = 9.04PTVFYY737 pKa = 10.71SINDD741 pKa = 3.51FEE743 pKa = 4.62TAVADD748 pKa = 4.2CASQASLLNFTKK760 pKa = 10.84ANLAGLTWYY769 pKa = 10.89VEE771 pKa = 4.2DD772 pKa = 4.0EE773 pKa = 4.59RR774 pKa = 11.84IVFDD778 pKa = 3.79SSADD782 pKa = 3.49TFVITTYY789 pKa = 11.41GDD791 pKa = 3.62DD792 pKa = 4.0GEE794 pKa = 5.61LGGGDD799 pKa = 4.66DD800 pKa = 3.57EE801 pKa = 4.79TFYY804 pKa = 10.8GTVADD809 pKa = 4.01YY810 pKa = 7.92ATNVIEE816 pKa = 4.86FSFSFTPGGDD826 pKa = 2.63IVGRR830 pKa = 11.84DD831 pKa = 4.15LIRR834 pKa = 11.84VKK836 pKa = 10.89AEE838 pKa = 3.95SSFQGKK844 pKa = 10.08VIFTAVLLWEE854 pKa = 4.86FYY856 pKa = 11.02DD857 pKa = 3.57WAEE860 pKa = 4.17SDD862 pKa = 4.14GDD864 pKa = 4.33NIDD867 pKa = 3.9KK868 pKa = 10.79AHH870 pKa = 6.4QGGEE874 pKa = 4.06LRR876 pKa = 11.84TAGYY880 pKa = 10.56AVDD883 pKa = 5.9ADD885 pKa = 4.29FDD887 pKa = 3.91WAAWQATPP895 pKa = 4.9
Molecular weight: 96.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2JKL8|A0A0S2JKL8_9GAMM EF-P post-translational modification enzyme B OS=Colwellia sp. MT41 OX=58049 GN=CMT41_03615 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.53RR3 pKa = 11.84TFQPSILKK11 pKa = 10.17RR12 pKa = 11.84KK13 pKa = 8.81RR14 pKa = 11.84NHH16 pKa = 5.4GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.25SGRR28 pKa = 11.84AVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.12GRR39 pKa = 11.84KK40 pKa = 8.87SLSAA44 pKa = 3.86
MM1 pKa = 7.45KK2 pKa = 9.53RR3 pKa = 11.84TFQPSILKK11 pKa = 10.17RR12 pKa = 11.84KK13 pKa = 8.81RR14 pKa = 11.84NHH16 pKa = 5.4GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.25SGRR28 pKa = 11.84AVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.12GRR39 pKa = 11.84KK40 pKa = 8.87SLSAA44 pKa = 3.86
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1168266 |
44 |
3376 |
334.7 |
37.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.661 ± 0.043 | 1.027 ± 0.016 |
5.521 ± 0.04 | 5.635 ± 0.035 |
4.28 ± 0.028 | 6.256 ± 0.04 |
2.23 ± 0.02 | 7.195 ± 0.032 |
6.153 ± 0.04 | 10.435 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.36 ± 0.021 | 4.887 ± 0.032 |
3.467 ± 0.022 | 4.796 ± 0.043 |
3.858 ± 0.032 | 6.967 ± 0.035 |
5.534 ± 0.032 | 6.451 ± 0.031 |
1.101 ± 0.013 | 3.185 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
