
Bacteroides phage crAss001
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae; crAss-like viruses
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
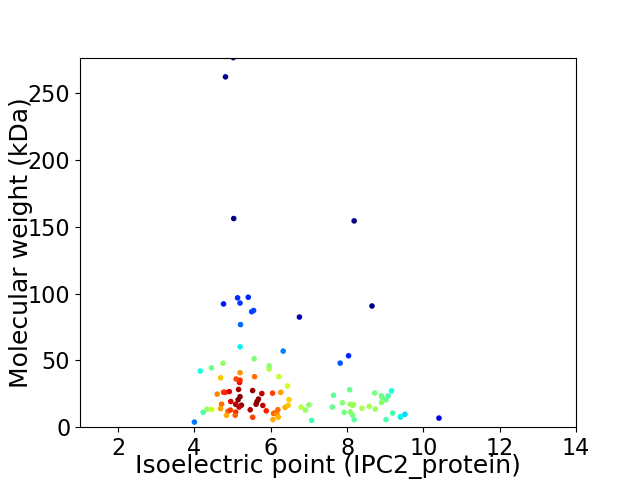
Virtual 2D-PAGE plot for 104 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
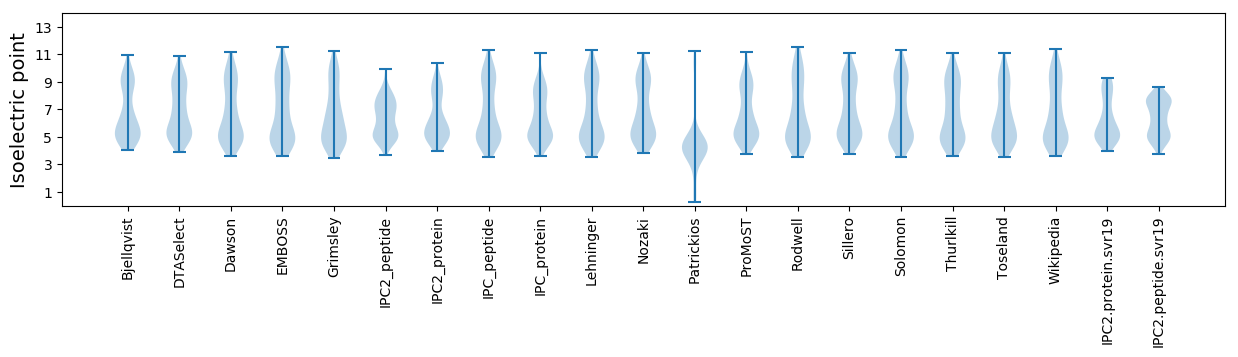
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385DVW5|A0A385DVW5_9CAUD Thiamine biosynthesis protein OS=Bacteroides phage crAss001 OX=2301731 GN=crAss001_76 PE=4 SV=1
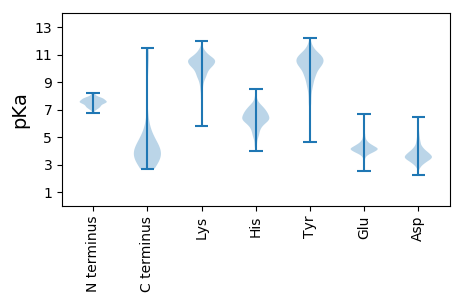
MM1 pKa = 7.7EE2 pKa = 6.01DD3 pKa = 3.66EE4 pKa = 4.45VLEE7 pKa = 4.24SAISQMTIDD16 pKa = 4.96AEE18 pKa = 4.59TAGLGHH24 pKa = 7.71PDD26 pKa = 4.18GIIIQTAIVGGSTYY40 pKa = 10.9APEE43 pKa = 4.33EE44 pKa = 3.79NLMYY48 pKa = 10.87SPASEE53 pKa = 4.32LNSAIALLQSGGWEE67 pKa = 4.09IIHH70 pKa = 6.42EE71 pKa = 4.26EE72 pKa = 3.99QHH74 pKa = 6.19VSGAYY79 pKa = 9.84LVTIGAVSMEE89 pKa = 4.27TPMLPVTLVVTLDD102 pKa = 3.83GSNLLHH108 pKa = 7.34DD109 pKa = 4.58AQNTEE114 pKa = 4.18DD115 pKa = 4.51APVEE119 pKa = 3.78IDD121 pKa = 3.61EE122 pKa = 4.29QGEE125 pKa = 4.21ALLEE129 pKa = 4.07AALAAEE135 pKa = 4.4EE136 pKa = 4.59VVIPPNSGSLLVDD149 pKa = 3.64EE150 pKa = 4.73ATSRR154 pKa = 11.84FSGAIWYY161 pKa = 9.21SAIQSKK167 pKa = 9.42TITLAGVGGIGSYY180 pKa = 10.75VGFLLARR187 pKa = 11.84LKK189 pKa = 10.66PAGLYY194 pKa = 10.5LYY196 pKa = 10.57DD197 pKa = 4.57PDD199 pKa = 4.54IVEE202 pKa = 4.15QANMSGQLYY211 pKa = 10.27GSQDD215 pKa = 3.11LGQAKK220 pKa = 9.91VSSLHH225 pKa = 7.28RR226 pKa = 11.84MLQVYY231 pKa = 10.18ANYY234 pKa = 9.22YY235 pKa = 10.36NSVAYY240 pKa = 8.15QEE242 pKa = 4.97RR243 pKa = 11.84FTDD246 pKa = 3.62EE247 pKa = 4.96SEE249 pKa = 4.03ATDD252 pKa = 4.01IMICGFDD259 pKa = 3.29NMEE262 pKa = 3.92ARR264 pKa = 11.84KK265 pKa = 9.99LFFDD269 pKa = 3.19KK270 pKa = 10.54WFEE273 pKa = 4.09HH274 pKa = 6.35VGNKK278 pKa = 9.84PEE280 pKa = 4.28GDD282 pKa = 3.09RR283 pKa = 11.84SKK285 pKa = 11.31CLFIDD290 pKa = 3.61GRR292 pKa = 11.84LAAEE296 pKa = 4.47EE297 pKa = 4.12FQVFAIQGNDD307 pKa = 2.47EE308 pKa = 4.13RR309 pKa = 11.84AIVEE313 pKa = 4.34YY314 pKa = 10.54KK315 pKa = 10.75NRR317 pKa = 11.84WLFSDD322 pKa = 4.39AVADD326 pKa = 3.99EE327 pKa = 5.06TICSYY332 pKa = 11.16KK333 pKa = 9.36QTTFMANMIASVMVNLFVNFVANEE357 pKa = 4.04CNPIIDD363 pKa = 3.65RR364 pKa = 11.84DD365 pKa = 4.16VPFMTQYY372 pKa = 11.26SADD375 pKa = 3.07TMYY378 pKa = 10.94FKK380 pKa = 11.42VEE382 pKa = 3.88MM383 pKa = 4.8
MM1 pKa = 7.7EE2 pKa = 6.01DD3 pKa = 3.66EE4 pKa = 4.45VLEE7 pKa = 4.24SAISQMTIDD16 pKa = 4.96AEE18 pKa = 4.59TAGLGHH24 pKa = 7.71PDD26 pKa = 4.18GIIIQTAIVGGSTYY40 pKa = 10.9APEE43 pKa = 4.33EE44 pKa = 3.79NLMYY48 pKa = 10.87SPASEE53 pKa = 4.32LNSAIALLQSGGWEE67 pKa = 4.09IIHH70 pKa = 6.42EE71 pKa = 4.26EE72 pKa = 3.99QHH74 pKa = 6.19VSGAYY79 pKa = 9.84LVTIGAVSMEE89 pKa = 4.27TPMLPVTLVVTLDD102 pKa = 3.83GSNLLHH108 pKa = 7.34DD109 pKa = 4.58AQNTEE114 pKa = 4.18DD115 pKa = 4.51APVEE119 pKa = 3.78IDD121 pKa = 3.61EE122 pKa = 4.29QGEE125 pKa = 4.21ALLEE129 pKa = 4.07AALAAEE135 pKa = 4.4EE136 pKa = 4.59VVIPPNSGSLLVDD149 pKa = 3.64EE150 pKa = 4.73ATSRR154 pKa = 11.84FSGAIWYY161 pKa = 9.21SAIQSKK167 pKa = 9.42TITLAGVGGIGSYY180 pKa = 10.75VGFLLARR187 pKa = 11.84LKK189 pKa = 10.66PAGLYY194 pKa = 10.5LYY196 pKa = 10.57DD197 pKa = 4.57PDD199 pKa = 4.54IVEE202 pKa = 4.15QANMSGQLYY211 pKa = 10.27GSQDD215 pKa = 3.11LGQAKK220 pKa = 9.91VSSLHH225 pKa = 7.28RR226 pKa = 11.84MLQVYY231 pKa = 10.18ANYY234 pKa = 9.22YY235 pKa = 10.36NSVAYY240 pKa = 8.15QEE242 pKa = 4.97RR243 pKa = 11.84FTDD246 pKa = 3.62EE247 pKa = 4.96SEE249 pKa = 4.03ATDD252 pKa = 4.01IMICGFDD259 pKa = 3.29NMEE262 pKa = 3.92ARR264 pKa = 11.84KK265 pKa = 9.99LFFDD269 pKa = 3.19KK270 pKa = 10.54WFEE273 pKa = 4.09HH274 pKa = 6.35VGNKK278 pKa = 9.84PEE280 pKa = 4.28GDD282 pKa = 3.09RR283 pKa = 11.84SKK285 pKa = 11.31CLFIDD290 pKa = 3.61GRR292 pKa = 11.84LAAEE296 pKa = 4.47EE297 pKa = 4.12FQVFAIQGNDD307 pKa = 2.47EE308 pKa = 4.13RR309 pKa = 11.84AIVEE313 pKa = 4.34YY314 pKa = 10.54KK315 pKa = 10.75NRR317 pKa = 11.84WLFSDD322 pKa = 4.39AVADD326 pKa = 3.99EE327 pKa = 5.06TICSYY332 pKa = 11.16KK333 pKa = 9.36QTTFMANMIASVMVNLFVNFVANEE357 pKa = 4.04CNPIIDD363 pKa = 3.65RR364 pKa = 11.84DD365 pKa = 4.16VPFMTQYY372 pKa = 11.26SADD375 pKa = 3.07TMYY378 pKa = 10.94FKK380 pKa = 11.42VEE382 pKa = 3.88MM383 pKa = 4.8
Molecular weight: 42.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385DTE1|A0A385DTE1_9CAUD Putative HNH endonuclease OS=Bacteroides phage crAss001 OX=2301731 GN=crAss001_8 PE=4 SV=1
MM1 pKa = 7.48SKK3 pKa = 10.79GRR5 pKa = 11.84VLTKK9 pKa = 10.5KK10 pKa = 10.18KK11 pKa = 8.98WGHH14 pKa = 5.44CARR17 pKa = 11.84YY18 pKa = 7.11WRR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 8.89KK23 pKa = 10.86YY24 pKa = 8.98RR25 pKa = 11.84TKK27 pKa = 10.59FPRR30 pKa = 11.84KK31 pKa = 9.19KK32 pKa = 10.31LNSFEE37 pKa = 4.2GTHH40 pKa = 6.42APIGYY45 pKa = 8.01PLKK48 pKa = 9.24YY49 pKa = 10.08LSWIMRR55 pKa = 3.81
MM1 pKa = 7.48SKK3 pKa = 10.79GRR5 pKa = 11.84VLTKK9 pKa = 10.5KK10 pKa = 10.18KK11 pKa = 8.98WGHH14 pKa = 5.44CARR17 pKa = 11.84YY18 pKa = 7.11WRR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 8.89KK23 pKa = 10.86YY24 pKa = 8.98RR25 pKa = 11.84TKK27 pKa = 10.59FPRR30 pKa = 11.84KK31 pKa = 9.19KK32 pKa = 10.31LNSFEE37 pKa = 4.2GTHH40 pKa = 6.42APIGYY45 pKa = 8.01PLKK48 pKa = 9.24YY49 pKa = 10.08LSWIMRR55 pKa = 3.81
Molecular weight: 6.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
31616 |
33 |
2412 |
304.0 |
34.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.902 ± 0.32 | 1.025 ± 0.148 |
6.041 ± 0.22 | 7.423 ± 0.305 |
4.213 ± 0.134 | 6.174 ± 0.27 |
1.215 ± 0.09 | 7.085 ± 0.242 |
7.695 ± 0.307 | 8.078 ± 0.18 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.562 ± 0.146 | 6.671 ± 0.291 |
3.318 ± 0.102 | 3.59 ± 0.195 |
4.438 ± 0.196 | 6.373 ± 0.199 |
5.807 ± 0.224 | 6.206 ± 0.18 |
1.253 ± 0.075 | 4.931 ± 0.208 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
