
Gigaspora margarita mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
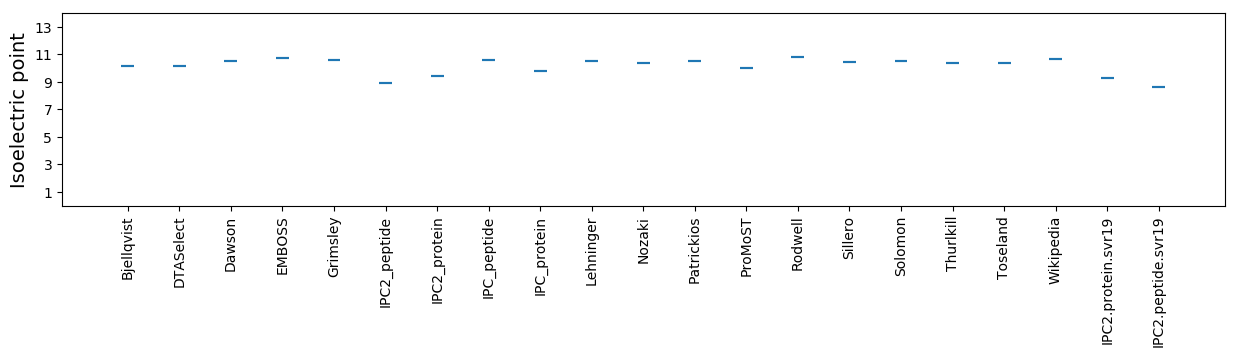
Average proteome isoelectric point is 9.26
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L0VZH3|A0A2L0VZH3_9VIRU RNA-dependent RNA polymerase OS=Gigaspora margarita mitovirus 1 OX=2082665 PE=4 SV=1
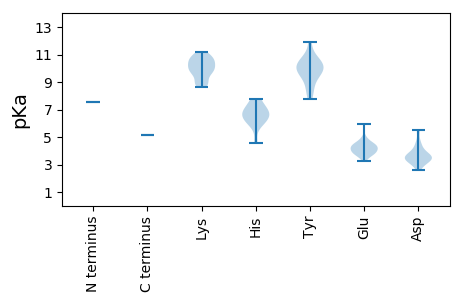
MM1 pKa = 7.56KK2 pKa = 10.45ALLLSLIRR10 pKa = 11.84VAKK13 pKa = 9.99EE14 pKa = 3.48RR15 pKa = 11.84VTVRR19 pKa = 11.84PVPLRR24 pKa = 11.84RR25 pKa = 11.84LNVKK29 pKa = 8.82VWHH32 pKa = 6.68HH33 pKa = 6.7LFPALVSVGRR43 pKa = 11.84LVIGRR48 pKa = 11.84ISRR51 pKa = 11.84DD52 pKa = 3.46RR53 pKa = 11.84IKK55 pKa = 11.15VLAFWAKK62 pKa = 9.74QCVAMIRR69 pKa = 11.84KK70 pKa = 8.98QGVKK74 pKa = 8.97GTCLWLKK81 pKa = 8.85TINVLIMASLPGSQLKK97 pKa = 9.72SQSRR101 pKa = 11.84AISKK105 pKa = 9.47VAVSVSRR112 pKa = 11.84DD113 pKa = 3.22GLPRR117 pKa = 11.84VIPKK121 pKa = 9.37QDD123 pKa = 2.75RR124 pKa = 11.84RR125 pKa = 11.84AIRR128 pKa = 11.84AGDD131 pKa = 3.64TNTIRR136 pKa = 11.84LWLSLSGMYY145 pKa = 9.51RR146 pKa = 11.84VLEE149 pKa = 4.27FPGKK153 pKa = 10.48AKK155 pKa = 10.36ISTITDD161 pKa = 4.1PGPRR165 pKa = 11.84ISPPLLSFFEE175 pKa = 4.47TFLKK179 pKa = 10.8RR180 pKa = 11.84EE181 pKa = 4.24FFRR184 pKa = 11.84NLGDD188 pKa = 3.61LTGLKK193 pKa = 8.83VTGIDD198 pKa = 3.63PAGLRR203 pKa = 11.84PTPLPLVTSAAGSWVVPSGKK223 pKa = 9.94YY224 pKa = 9.84DD225 pKa = 3.58LRR227 pKa = 11.84VSSLGTAAPAAWNWVNGHH245 pKa = 5.95WGEE248 pKa = 3.87ALLRR252 pKa = 11.84YY253 pKa = 9.3SQAIGSWDD261 pKa = 3.69TTNSWYY267 pKa = 10.74RR268 pKa = 11.84LVEE271 pKa = 3.72EE272 pKa = 4.36SARR275 pKa = 11.84YY276 pKa = 9.42SHH278 pKa = 6.99FCSRR282 pKa = 11.84MSRR285 pKa = 11.84GRR287 pKa = 11.84ISFKK291 pKa = 10.71DD292 pKa = 3.76EE293 pKa = 3.63PAGKK297 pKa = 9.74VRR299 pKa = 11.84AFAMVDD305 pKa = 3.06YY306 pKa = 7.77WTQCSLKK313 pKa = 9.72PLHH316 pKa = 7.13DD317 pKa = 4.42LLFSILKK324 pKa = 8.75EE325 pKa = 3.96IPQDD329 pKa = 3.53GTFDD333 pKa = 3.29QMAPAKK339 pKa = 9.66EE340 pKa = 3.93LLSVRR345 pKa = 11.84RR346 pKa = 11.84LATEE350 pKa = 4.13TWWSLDD356 pKa = 3.19LSAATDD362 pKa = 4.08RR363 pKa = 11.84FPLALQKK370 pKa = 11.0LAMKK374 pKa = 10.04YY375 pKa = 10.03LVSEE379 pKa = 4.94EE380 pKa = 4.36YY381 pKa = 10.42AAAWATLMVDD391 pKa = 3.68RR392 pKa = 11.84PFRR395 pKa = 11.84LPVGYY400 pKa = 10.1KK401 pKa = 10.25PRR403 pKa = 11.84DD404 pKa = 3.3CRR406 pKa = 11.84YY407 pKa = 10.46AVGQPMGAYY416 pKa = 10.02SSWAAFALTHH426 pKa = 6.24HH427 pKa = 6.52ATVQFAARR435 pKa = 11.84LSGLSGWFRR444 pKa = 11.84DD445 pKa = 3.91YY446 pKa = 11.88ALLGDD451 pKa = 5.49DD452 pKa = 5.37IIIANDD458 pKa = 3.46RR459 pKa = 11.84VAHH462 pKa = 6.7KK463 pKa = 10.36YY464 pKa = 9.75RR465 pKa = 11.84WLLSQLGVEE474 pKa = 4.48CSLAKK479 pKa = 10.68SMASNQRR486 pKa = 11.84TFEE489 pKa = 3.79FAKK492 pKa = 9.91RR493 pKa = 11.84VLFRR497 pKa = 11.84GVDD500 pKa = 3.4VSGFPWKK507 pKa = 10.3LWRR510 pKa = 11.84IGQRR514 pKa = 11.84SLAAMISLAQRR525 pKa = 11.84VSFGTAPLNLASLVKK540 pKa = 10.69ALGAGMKK547 pKa = 8.64ATSRR551 pKa = 11.84AYY553 pKa = 8.52TAWRR557 pKa = 11.84HH558 pKa = 4.56MPRR561 pKa = 11.84GLKK564 pKa = 10.22ALLIILTHH572 pKa = 5.99PQANTFLSRR581 pKa = 11.84QTWLDD586 pKa = 3.1WLTAPGPVLPSGYY599 pKa = 10.4KK600 pKa = 9.94VDD602 pKa = 3.42TSTWLTPWLSGFRR615 pKa = 11.84EE616 pKa = 4.54EE617 pKa = 4.64YY618 pKa = 10.82LNPALEE624 pKa = 4.1FLEE627 pKa = 4.63EE628 pKa = 4.31EE629 pKa = 4.26STLDD633 pKa = 4.6LFEE636 pKa = 5.98PDD638 pKa = 2.61IDD640 pKa = 4.62RR641 pKa = 11.84RR642 pKa = 11.84DD643 pKa = 3.56AQGNLTRR650 pKa = 11.84DD651 pKa = 3.32SHH653 pKa = 7.64YY654 pKa = 8.47MAEE657 pKa = 4.97FIRR660 pKa = 11.84FRR662 pKa = 11.84RR663 pKa = 11.84AYY665 pKa = 10.04LFSKK669 pKa = 10.74LEE671 pKa = 3.99SLVSVEE677 pKa = 3.7RR678 pKa = 11.84SKK680 pKa = 11.03KK681 pKa = 9.26IRR683 pKa = 11.84QLEE686 pKa = 3.81EE687 pKa = 3.26RR688 pKa = 11.84AAKK691 pKa = 10.15IEE693 pKa = 4.08ADD695 pKa = 3.55LAHH698 pKa = 6.3LQKK701 pKa = 11.08LSISLQARR709 pKa = 11.84NVSGMFNSMMKK720 pKa = 10.41RR721 pKa = 11.84LEE723 pKa = 3.92EE724 pKa = 4.4SIAAVPLPLPWLVWARR740 pKa = 11.84EE741 pKa = 4.09EE742 pKa = 4.4NEE744 pKa = 3.65NRR746 pKa = 11.84RR747 pKa = 11.84PATVLVSLWEE757 pKa = 3.88RR758 pKa = 11.84WRR760 pKa = 11.84LRR762 pKa = 11.84IHH764 pKa = 7.77RR765 pKa = 11.84SRR767 pKa = 11.84KK768 pKa = 9.8LPTHH772 pKa = 7.07ANPQGEE778 pKa = 4.22AVKK781 pKa = 10.79ARR783 pKa = 11.84FEE785 pKa = 4.65KK786 pKa = 10.71PFSTDD791 pKa = 3.09FDD793 pKa = 4.06EE794 pKa = 5.12
MM1 pKa = 7.56KK2 pKa = 10.45ALLLSLIRR10 pKa = 11.84VAKK13 pKa = 9.99EE14 pKa = 3.48RR15 pKa = 11.84VTVRR19 pKa = 11.84PVPLRR24 pKa = 11.84RR25 pKa = 11.84LNVKK29 pKa = 8.82VWHH32 pKa = 6.68HH33 pKa = 6.7LFPALVSVGRR43 pKa = 11.84LVIGRR48 pKa = 11.84ISRR51 pKa = 11.84DD52 pKa = 3.46RR53 pKa = 11.84IKK55 pKa = 11.15VLAFWAKK62 pKa = 9.74QCVAMIRR69 pKa = 11.84KK70 pKa = 8.98QGVKK74 pKa = 8.97GTCLWLKK81 pKa = 8.85TINVLIMASLPGSQLKK97 pKa = 9.72SQSRR101 pKa = 11.84AISKK105 pKa = 9.47VAVSVSRR112 pKa = 11.84DD113 pKa = 3.22GLPRR117 pKa = 11.84VIPKK121 pKa = 9.37QDD123 pKa = 2.75RR124 pKa = 11.84RR125 pKa = 11.84AIRR128 pKa = 11.84AGDD131 pKa = 3.64TNTIRR136 pKa = 11.84LWLSLSGMYY145 pKa = 9.51RR146 pKa = 11.84VLEE149 pKa = 4.27FPGKK153 pKa = 10.48AKK155 pKa = 10.36ISTITDD161 pKa = 4.1PGPRR165 pKa = 11.84ISPPLLSFFEE175 pKa = 4.47TFLKK179 pKa = 10.8RR180 pKa = 11.84EE181 pKa = 4.24FFRR184 pKa = 11.84NLGDD188 pKa = 3.61LTGLKK193 pKa = 8.83VTGIDD198 pKa = 3.63PAGLRR203 pKa = 11.84PTPLPLVTSAAGSWVVPSGKK223 pKa = 9.94YY224 pKa = 9.84DD225 pKa = 3.58LRR227 pKa = 11.84VSSLGTAAPAAWNWVNGHH245 pKa = 5.95WGEE248 pKa = 3.87ALLRR252 pKa = 11.84YY253 pKa = 9.3SQAIGSWDD261 pKa = 3.69TTNSWYY267 pKa = 10.74RR268 pKa = 11.84LVEE271 pKa = 3.72EE272 pKa = 4.36SARR275 pKa = 11.84YY276 pKa = 9.42SHH278 pKa = 6.99FCSRR282 pKa = 11.84MSRR285 pKa = 11.84GRR287 pKa = 11.84ISFKK291 pKa = 10.71DD292 pKa = 3.76EE293 pKa = 3.63PAGKK297 pKa = 9.74VRR299 pKa = 11.84AFAMVDD305 pKa = 3.06YY306 pKa = 7.77WTQCSLKK313 pKa = 9.72PLHH316 pKa = 7.13DD317 pKa = 4.42LLFSILKK324 pKa = 8.75EE325 pKa = 3.96IPQDD329 pKa = 3.53GTFDD333 pKa = 3.29QMAPAKK339 pKa = 9.66EE340 pKa = 3.93LLSVRR345 pKa = 11.84RR346 pKa = 11.84LATEE350 pKa = 4.13TWWSLDD356 pKa = 3.19LSAATDD362 pKa = 4.08RR363 pKa = 11.84FPLALQKK370 pKa = 11.0LAMKK374 pKa = 10.04YY375 pKa = 10.03LVSEE379 pKa = 4.94EE380 pKa = 4.36YY381 pKa = 10.42AAAWATLMVDD391 pKa = 3.68RR392 pKa = 11.84PFRR395 pKa = 11.84LPVGYY400 pKa = 10.1KK401 pKa = 10.25PRR403 pKa = 11.84DD404 pKa = 3.3CRR406 pKa = 11.84YY407 pKa = 10.46AVGQPMGAYY416 pKa = 10.02SSWAAFALTHH426 pKa = 6.24HH427 pKa = 6.52ATVQFAARR435 pKa = 11.84LSGLSGWFRR444 pKa = 11.84DD445 pKa = 3.91YY446 pKa = 11.88ALLGDD451 pKa = 5.49DD452 pKa = 5.37IIIANDD458 pKa = 3.46RR459 pKa = 11.84VAHH462 pKa = 6.7KK463 pKa = 10.36YY464 pKa = 9.75RR465 pKa = 11.84WLLSQLGVEE474 pKa = 4.48CSLAKK479 pKa = 10.68SMASNQRR486 pKa = 11.84TFEE489 pKa = 3.79FAKK492 pKa = 9.91RR493 pKa = 11.84VLFRR497 pKa = 11.84GVDD500 pKa = 3.4VSGFPWKK507 pKa = 10.3LWRR510 pKa = 11.84IGQRR514 pKa = 11.84SLAAMISLAQRR525 pKa = 11.84VSFGTAPLNLASLVKK540 pKa = 10.69ALGAGMKK547 pKa = 8.64ATSRR551 pKa = 11.84AYY553 pKa = 8.52TAWRR557 pKa = 11.84HH558 pKa = 4.56MPRR561 pKa = 11.84GLKK564 pKa = 10.22ALLIILTHH572 pKa = 5.99PQANTFLSRR581 pKa = 11.84QTWLDD586 pKa = 3.1WLTAPGPVLPSGYY599 pKa = 10.4KK600 pKa = 9.94VDD602 pKa = 3.42TSTWLTPWLSGFRR615 pKa = 11.84EE616 pKa = 4.54EE617 pKa = 4.64YY618 pKa = 10.82LNPALEE624 pKa = 4.1FLEE627 pKa = 4.63EE628 pKa = 4.31EE629 pKa = 4.26STLDD633 pKa = 4.6LFEE636 pKa = 5.98PDD638 pKa = 2.61IDD640 pKa = 4.62RR641 pKa = 11.84RR642 pKa = 11.84DD643 pKa = 3.56AQGNLTRR650 pKa = 11.84DD651 pKa = 3.32SHH653 pKa = 7.64YY654 pKa = 8.47MAEE657 pKa = 4.97FIRR660 pKa = 11.84FRR662 pKa = 11.84RR663 pKa = 11.84AYY665 pKa = 10.04LFSKK669 pKa = 10.74LEE671 pKa = 3.99SLVSVEE677 pKa = 3.7RR678 pKa = 11.84SKK680 pKa = 11.03KK681 pKa = 9.26IRR683 pKa = 11.84QLEE686 pKa = 3.81EE687 pKa = 3.26RR688 pKa = 11.84AAKK691 pKa = 10.15IEE693 pKa = 4.08ADD695 pKa = 3.55LAHH698 pKa = 6.3LQKK701 pKa = 11.08LSISLQARR709 pKa = 11.84NVSGMFNSMMKK720 pKa = 10.41RR721 pKa = 11.84LEE723 pKa = 3.92EE724 pKa = 4.4SIAAVPLPLPWLVWARR740 pKa = 11.84EE741 pKa = 4.09EE742 pKa = 4.4NEE744 pKa = 3.65NRR746 pKa = 11.84RR747 pKa = 11.84PATVLVSLWEE757 pKa = 3.88RR758 pKa = 11.84WRR760 pKa = 11.84LRR762 pKa = 11.84IHH764 pKa = 7.77RR765 pKa = 11.84SRR767 pKa = 11.84KK768 pKa = 9.8LPTHH772 pKa = 7.07ANPQGEE778 pKa = 4.22AVKK781 pKa = 10.79ARR783 pKa = 11.84FEE785 pKa = 4.65KK786 pKa = 10.71PFSTDD791 pKa = 3.09FDD793 pKa = 4.06EE794 pKa = 5.12
Molecular weight: 89.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L0VZH3|A0A2L0VZH3_9VIRU RNA-dependent RNA polymerase OS=Gigaspora margarita mitovirus 1 OX=2082665 PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 10.45ALLLSLIRR10 pKa = 11.84VAKK13 pKa = 9.99EE14 pKa = 3.48RR15 pKa = 11.84VTVRR19 pKa = 11.84PVPLRR24 pKa = 11.84RR25 pKa = 11.84LNVKK29 pKa = 8.82VWHH32 pKa = 6.68HH33 pKa = 6.7LFPALVSVGRR43 pKa = 11.84LVIGRR48 pKa = 11.84ISRR51 pKa = 11.84DD52 pKa = 3.46RR53 pKa = 11.84IKK55 pKa = 11.15VLAFWAKK62 pKa = 9.74QCVAMIRR69 pKa = 11.84KK70 pKa = 8.98QGVKK74 pKa = 8.97GTCLWLKK81 pKa = 8.85TINVLIMASLPGSQLKK97 pKa = 9.72SQSRR101 pKa = 11.84AISKK105 pKa = 9.47VAVSVSRR112 pKa = 11.84DD113 pKa = 3.22GLPRR117 pKa = 11.84VIPKK121 pKa = 9.37QDD123 pKa = 2.75RR124 pKa = 11.84RR125 pKa = 11.84AIRR128 pKa = 11.84AGDD131 pKa = 3.64TNTIRR136 pKa = 11.84LWLSLSGMYY145 pKa = 9.51RR146 pKa = 11.84VLEE149 pKa = 4.27FPGKK153 pKa = 10.48AKK155 pKa = 10.36ISTITDD161 pKa = 4.1PGPRR165 pKa = 11.84ISPPLLSFFEE175 pKa = 4.47TFLKK179 pKa = 10.8RR180 pKa = 11.84EE181 pKa = 4.24FFRR184 pKa = 11.84NLGDD188 pKa = 3.61LTGLKK193 pKa = 8.83VTGIDD198 pKa = 3.63PAGLRR203 pKa = 11.84PTPLPLVTSAAGSWVVPSGKK223 pKa = 9.94YY224 pKa = 9.84DD225 pKa = 3.58LRR227 pKa = 11.84VSSLGTAAPAAWNWVNGHH245 pKa = 5.95WGEE248 pKa = 3.87ALLRR252 pKa = 11.84YY253 pKa = 9.3SQAIGSWDD261 pKa = 3.69TTNSWYY267 pKa = 10.74RR268 pKa = 11.84LVEE271 pKa = 3.72EE272 pKa = 4.36SARR275 pKa = 11.84YY276 pKa = 9.42SHH278 pKa = 6.99FCSRR282 pKa = 11.84MSRR285 pKa = 11.84GRR287 pKa = 11.84ISFKK291 pKa = 10.71DD292 pKa = 3.76EE293 pKa = 3.63PAGKK297 pKa = 9.74VRR299 pKa = 11.84AFAMVDD305 pKa = 3.06YY306 pKa = 7.77WTQCSLKK313 pKa = 9.72PLHH316 pKa = 7.13DD317 pKa = 4.42LLFSILKK324 pKa = 8.75EE325 pKa = 3.96IPQDD329 pKa = 3.53GTFDD333 pKa = 3.29QMAPAKK339 pKa = 9.66EE340 pKa = 3.93LLSVRR345 pKa = 11.84RR346 pKa = 11.84LATEE350 pKa = 4.13TWWSLDD356 pKa = 3.19LSAATDD362 pKa = 4.08RR363 pKa = 11.84FPLALQKK370 pKa = 11.0LAMKK374 pKa = 10.04YY375 pKa = 10.03LVSEE379 pKa = 4.94EE380 pKa = 4.36YY381 pKa = 10.42AAAWATLMVDD391 pKa = 3.68RR392 pKa = 11.84PFRR395 pKa = 11.84LPVGYY400 pKa = 10.1KK401 pKa = 10.25PRR403 pKa = 11.84DD404 pKa = 3.3CRR406 pKa = 11.84YY407 pKa = 10.46AVGQPMGAYY416 pKa = 10.02SSWAAFALTHH426 pKa = 6.24HH427 pKa = 6.52ATVQFAARR435 pKa = 11.84LSGLSGWFRR444 pKa = 11.84DD445 pKa = 3.91YY446 pKa = 11.88ALLGDD451 pKa = 5.49DD452 pKa = 5.37IIIANDD458 pKa = 3.46RR459 pKa = 11.84VAHH462 pKa = 6.7KK463 pKa = 10.36YY464 pKa = 9.75RR465 pKa = 11.84WLLSQLGVEE474 pKa = 4.48CSLAKK479 pKa = 10.68SMASNQRR486 pKa = 11.84TFEE489 pKa = 3.79FAKK492 pKa = 9.91RR493 pKa = 11.84VLFRR497 pKa = 11.84GVDD500 pKa = 3.4VSGFPWKK507 pKa = 10.3LWRR510 pKa = 11.84IGQRR514 pKa = 11.84SLAAMISLAQRR525 pKa = 11.84VSFGTAPLNLASLVKK540 pKa = 10.69ALGAGMKK547 pKa = 8.64ATSRR551 pKa = 11.84AYY553 pKa = 8.52TAWRR557 pKa = 11.84HH558 pKa = 4.56MPRR561 pKa = 11.84GLKK564 pKa = 10.22ALLIILTHH572 pKa = 5.99PQANTFLSRR581 pKa = 11.84QTWLDD586 pKa = 3.1WLTAPGPVLPSGYY599 pKa = 10.4KK600 pKa = 9.94VDD602 pKa = 3.42TSTWLTPWLSGFRR615 pKa = 11.84EE616 pKa = 4.54EE617 pKa = 4.64YY618 pKa = 10.82LNPALEE624 pKa = 4.1FLEE627 pKa = 4.63EE628 pKa = 4.31EE629 pKa = 4.26STLDD633 pKa = 4.6LFEE636 pKa = 5.98PDD638 pKa = 2.61IDD640 pKa = 4.62RR641 pKa = 11.84RR642 pKa = 11.84DD643 pKa = 3.56AQGNLTRR650 pKa = 11.84DD651 pKa = 3.32SHH653 pKa = 7.64YY654 pKa = 8.47MAEE657 pKa = 4.97FIRR660 pKa = 11.84FRR662 pKa = 11.84RR663 pKa = 11.84AYY665 pKa = 10.04LFSKK669 pKa = 10.74LEE671 pKa = 3.99SLVSVEE677 pKa = 3.7RR678 pKa = 11.84SKK680 pKa = 11.03KK681 pKa = 9.26IRR683 pKa = 11.84QLEE686 pKa = 3.81EE687 pKa = 3.26RR688 pKa = 11.84AAKK691 pKa = 10.15IEE693 pKa = 4.08ADD695 pKa = 3.55LAHH698 pKa = 6.3LQKK701 pKa = 11.08LSISLQARR709 pKa = 11.84NVSGMFNSMMKK720 pKa = 10.41RR721 pKa = 11.84LEE723 pKa = 3.92EE724 pKa = 4.4SIAAVPLPLPWLVWARR740 pKa = 11.84EE741 pKa = 4.09EE742 pKa = 4.4NEE744 pKa = 3.65NRR746 pKa = 11.84RR747 pKa = 11.84PATVLVSLWEE757 pKa = 3.88RR758 pKa = 11.84WRR760 pKa = 11.84LRR762 pKa = 11.84IHH764 pKa = 7.77RR765 pKa = 11.84SRR767 pKa = 11.84KK768 pKa = 9.8LPTHH772 pKa = 7.07ANPQGEE778 pKa = 4.22AVKK781 pKa = 10.79ARR783 pKa = 11.84FEE785 pKa = 4.65KK786 pKa = 10.71PFSTDD791 pKa = 3.09FDD793 pKa = 4.06EE794 pKa = 5.12
MM1 pKa = 7.56KK2 pKa = 10.45ALLLSLIRR10 pKa = 11.84VAKK13 pKa = 9.99EE14 pKa = 3.48RR15 pKa = 11.84VTVRR19 pKa = 11.84PVPLRR24 pKa = 11.84RR25 pKa = 11.84LNVKK29 pKa = 8.82VWHH32 pKa = 6.68HH33 pKa = 6.7LFPALVSVGRR43 pKa = 11.84LVIGRR48 pKa = 11.84ISRR51 pKa = 11.84DD52 pKa = 3.46RR53 pKa = 11.84IKK55 pKa = 11.15VLAFWAKK62 pKa = 9.74QCVAMIRR69 pKa = 11.84KK70 pKa = 8.98QGVKK74 pKa = 8.97GTCLWLKK81 pKa = 8.85TINVLIMASLPGSQLKK97 pKa = 9.72SQSRR101 pKa = 11.84AISKK105 pKa = 9.47VAVSVSRR112 pKa = 11.84DD113 pKa = 3.22GLPRR117 pKa = 11.84VIPKK121 pKa = 9.37QDD123 pKa = 2.75RR124 pKa = 11.84RR125 pKa = 11.84AIRR128 pKa = 11.84AGDD131 pKa = 3.64TNTIRR136 pKa = 11.84LWLSLSGMYY145 pKa = 9.51RR146 pKa = 11.84VLEE149 pKa = 4.27FPGKK153 pKa = 10.48AKK155 pKa = 10.36ISTITDD161 pKa = 4.1PGPRR165 pKa = 11.84ISPPLLSFFEE175 pKa = 4.47TFLKK179 pKa = 10.8RR180 pKa = 11.84EE181 pKa = 4.24FFRR184 pKa = 11.84NLGDD188 pKa = 3.61LTGLKK193 pKa = 8.83VTGIDD198 pKa = 3.63PAGLRR203 pKa = 11.84PTPLPLVTSAAGSWVVPSGKK223 pKa = 9.94YY224 pKa = 9.84DD225 pKa = 3.58LRR227 pKa = 11.84VSSLGTAAPAAWNWVNGHH245 pKa = 5.95WGEE248 pKa = 3.87ALLRR252 pKa = 11.84YY253 pKa = 9.3SQAIGSWDD261 pKa = 3.69TTNSWYY267 pKa = 10.74RR268 pKa = 11.84LVEE271 pKa = 3.72EE272 pKa = 4.36SARR275 pKa = 11.84YY276 pKa = 9.42SHH278 pKa = 6.99FCSRR282 pKa = 11.84MSRR285 pKa = 11.84GRR287 pKa = 11.84ISFKK291 pKa = 10.71DD292 pKa = 3.76EE293 pKa = 3.63PAGKK297 pKa = 9.74VRR299 pKa = 11.84AFAMVDD305 pKa = 3.06YY306 pKa = 7.77WTQCSLKK313 pKa = 9.72PLHH316 pKa = 7.13DD317 pKa = 4.42LLFSILKK324 pKa = 8.75EE325 pKa = 3.96IPQDD329 pKa = 3.53GTFDD333 pKa = 3.29QMAPAKK339 pKa = 9.66EE340 pKa = 3.93LLSVRR345 pKa = 11.84RR346 pKa = 11.84LATEE350 pKa = 4.13TWWSLDD356 pKa = 3.19LSAATDD362 pKa = 4.08RR363 pKa = 11.84FPLALQKK370 pKa = 11.0LAMKK374 pKa = 10.04YY375 pKa = 10.03LVSEE379 pKa = 4.94EE380 pKa = 4.36YY381 pKa = 10.42AAAWATLMVDD391 pKa = 3.68RR392 pKa = 11.84PFRR395 pKa = 11.84LPVGYY400 pKa = 10.1KK401 pKa = 10.25PRR403 pKa = 11.84DD404 pKa = 3.3CRR406 pKa = 11.84YY407 pKa = 10.46AVGQPMGAYY416 pKa = 10.02SSWAAFALTHH426 pKa = 6.24HH427 pKa = 6.52ATVQFAARR435 pKa = 11.84LSGLSGWFRR444 pKa = 11.84DD445 pKa = 3.91YY446 pKa = 11.88ALLGDD451 pKa = 5.49DD452 pKa = 5.37IIIANDD458 pKa = 3.46RR459 pKa = 11.84VAHH462 pKa = 6.7KK463 pKa = 10.36YY464 pKa = 9.75RR465 pKa = 11.84WLLSQLGVEE474 pKa = 4.48CSLAKK479 pKa = 10.68SMASNQRR486 pKa = 11.84TFEE489 pKa = 3.79FAKK492 pKa = 9.91RR493 pKa = 11.84VLFRR497 pKa = 11.84GVDD500 pKa = 3.4VSGFPWKK507 pKa = 10.3LWRR510 pKa = 11.84IGQRR514 pKa = 11.84SLAAMISLAQRR525 pKa = 11.84VSFGTAPLNLASLVKK540 pKa = 10.69ALGAGMKK547 pKa = 8.64ATSRR551 pKa = 11.84AYY553 pKa = 8.52TAWRR557 pKa = 11.84HH558 pKa = 4.56MPRR561 pKa = 11.84GLKK564 pKa = 10.22ALLIILTHH572 pKa = 5.99PQANTFLSRR581 pKa = 11.84QTWLDD586 pKa = 3.1WLTAPGPVLPSGYY599 pKa = 10.4KK600 pKa = 9.94VDD602 pKa = 3.42TSTWLTPWLSGFRR615 pKa = 11.84EE616 pKa = 4.54EE617 pKa = 4.64YY618 pKa = 10.82LNPALEE624 pKa = 4.1FLEE627 pKa = 4.63EE628 pKa = 4.31EE629 pKa = 4.26STLDD633 pKa = 4.6LFEE636 pKa = 5.98PDD638 pKa = 2.61IDD640 pKa = 4.62RR641 pKa = 11.84RR642 pKa = 11.84DD643 pKa = 3.56AQGNLTRR650 pKa = 11.84DD651 pKa = 3.32SHH653 pKa = 7.64YY654 pKa = 8.47MAEE657 pKa = 4.97FIRR660 pKa = 11.84FRR662 pKa = 11.84RR663 pKa = 11.84AYY665 pKa = 10.04LFSKK669 pKa = 10.74LEE671 pKa = 3.99SLVSVEE677 pKa = 3.7RR678 pKa = 11.84SKK680 pKa = 11.03KK681 pKa = 9.26IRR683 pKa = 11.84QLEE686 pKa = 3.81EE687 pKa = 3.26RR688 pKa = 11.84AAKK691 pKa = 10.15IEE693 pKa = 4.08ADD695 pKa = 3.55LAHH698 pKa = 6.3LQKK701 pKa = 11.08LSISLQARR709 pKa = 11.84NVSGMFNSMMKK720 pKa = 10.41RR721 pKa = 11.84LEE723 pKa = 3.92EE724 pKa = 4.4SIAAVPLPLPWLVWARR740 pKa = 11.84EE741 pKa = 4.09EE742 pKa = 4.4NEE744 pKa = 3.65NRR746 pKa = 11.84RR747 pKa = 11.84PATVLVSLWEE757 pKa = 3.88RR758 pKa = 11.84WRR760 pKa = 11.84LRR762 pKa = 11.84IHH764 pKa = 7.77RR765 pKa = 11.84SRR767 pKa = 11.84KK768 pKa = 9.8LPTHH772 pKa = 7.07ANPQGEE778 pKa = 4.22AVKK781 pKa = 10.79ARR783 pKa = 11.84FEE785 pKa = 4.65KK786 pKa = 10.71PFSTDD791 pKa = 3.09FDD793 pKa = 4.06EE794 pKa = 5.12
Molecular weight: 89.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
794 |
794 |
794 |
794.0 |
89.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.95 ± 0.0 | 0.756 ± 0.0 |
4.156 ± 0.0 | 4.66 ± 0.0 |
4.282 ± 0.0 | 5.416 ± 0.0 |
1.763 ± 0.0 | 4.156 ± 0.0 |
5.164 ± 0.0 | 12.469 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.267 ± 0.0 | 2.267 ± 0.0 |
5.416 ± 0.0 | 2.897 ± 0.0 |
9.068 ± 0.0 | 8.312 ± 0.0 |
4.912 ± 0.0 | 6.297 ± 0.0 |
3.526 ± 0.0 | 2.267 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
