
Aurantimonas manganoxydans (strain ATCC BAA-1229 / DSM 21871 / SI85-9A1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Aurantimonas; Aurantimonas manganoxydans
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
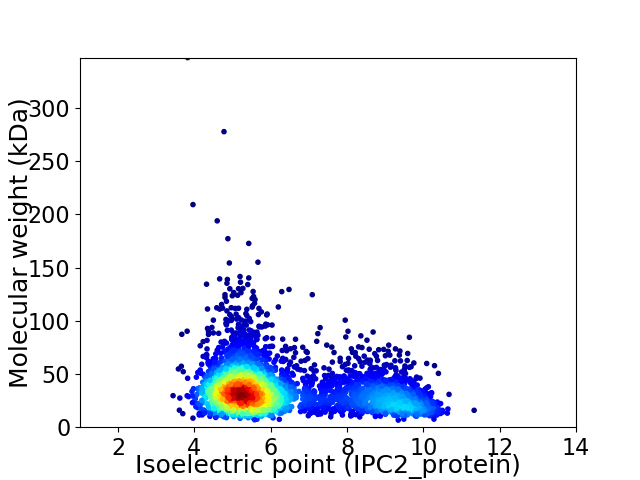
Virtual 2D-PAGE plot for 3625 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1YEJ7|Q1YEJ7_AURMS Putative exonuclease OS=Aurantimonas manganoxydans (strain ATCC BAA-1229 / DSM 21871 / SI85-9A1) OX=287752 GN=SI859A1_01078 PE=4 SV=1
MM1 pKa = 7.85TGGQDD6 pKa = 3.5MNIKK10 pKa = 10.36SLLLGSAAALVAVSGARR27 pKa = 11.84AADD30 pKa = 3.68VIMPVAPEE38 pKa = 3.71PVDD41 pKa = 3.52YY42 pKa = 11.42VRR44 pKa = 11.84VCDD47 pKa = 3.88VYY49 pKa = 10.81GTGFYY54 pKa = 10.01YY55 pKa = 9.98IPGTEE60 pKa = 3.92TCLSINGYY68 pKa = 8.74VRR70 pKa = 11.84FQYY73 pKa = 10.71NVAGGIEE80 pKa = 4.43DD81 pKa = 5.31DD82 pKa = 4.58FGNDD86 pKa = 4.01LLTNSDD92 pKa = 3.37GDD94 pKa = 3.77NYY96 pKa = 10.59QAGSVVRR103 pKa = 11.84VRR105 pKa = 11.84LNFDD109 pKa = 2.97ARR111 pKa = 11.84EE112 pKa = 3.94EE113 pKa = 4.57TEE115 pKa = 4.58LGTLRR120 pKa = 11.84AYY122 pKa = 10.59ARR124 pKa = 11.84VQAQNAGGAYY134 pKa = 10.65SNDD137 pKa = 2.71ASYY140 pKa = 11.86NMDD143 pKa = 3.2MGFIQLGGLTMGYY156 pKa = 10.52LDD158 pKa = 4.17TLWTEE163 pKa = 4.38GDD165 pKa = 3.77GLLTDD170 pKa = 3.6TDD172 pKa = 4.77LPVGDD177 pKa = 4.24YY178 pKa = 7.2QTNRR182 pKa = 11.84ISYY185 pKa = 8.18TYY187 pKa = 10.27AANGFTAALSLEE199 pKa = 4.47DD200 pKa = 4.45DD201 pKa = 3.67ATGNFAPDD209 pKa = 3.32VVGRR213 pKa = 11.84LAYY216 pKa = 10.19AGGWGSVYY224 pKa = 10.65LAGSYY229 pKa = 10.52DD230 pKa = 3.68EE231 pKa = 4.99QVDD234 pKa = 3.96GRR236 pKa = 11.84LLTPASFTAAAASGLQVNSAGVIVPASATFVSGIDD271 pKa = 3.34PTTDD275 pKa = 3.08DD276 pKa = 4.15GAFALKK282 pKa = 10.53AGLSLTDD289 pKa = 5.44LIAADD294 pKa = 4.24SEE296 pKa = 4.54LKK298 pKa = 10.46IEE300 pKa = 4.32GSYY303 pKa = 11.35AFDD306 pKa = 4.1PSSYY310 pKa = 9.69STIGLFSNRR319 pKa = 11.84ATLGTQLDD327 pKa = 4.97LLTGTLPVEE336 pKa = 4.11WQIGAGYY343 pKa = 9.37AQQFGKK349 pKa = 10.79LGVAVSGVYY358 pKa = 10.82GEE360 pKa = 4.52TFDD363 pKa = 3.68TYY365 pKa = 10.95RR366 pKa = 11.84YY367 pKa = 8.03TNATTFSNRR376 pKa = 11.84GDD378 pKa = 3.37GEE380 pKa = 4.46YY381 pKa = 10.82YY382 pKa = 10.8KK383 pKa = 11.07LVGNVGYY390 pKa = 10.47EE391 pKa = 3.6ITNNFDD397 pKa = 3.11MLAEE401 pKa = 4.15ISYY404 pKa = 11.49ANIDD408 pKa = 3.46FDD410 pKa = 4.09GQNGFVDD417 pKa = 3.57NSLDD421 pKa = 3.41QTAGFLRR428 pKa = 11.84FVRR431 pKa = 11.84SFF433 pKa = 3.04
MM1 pKa = 7.85TGGQDD6 pKa = 3.5MNIKK10 pKa = 10.36SLLLGSAAALVAVSGARR27 pKa = 11.84AADD30 pKa = 3.68VIMPVAPEE38 pKa = 3.71PVDD41 pKa = 3.52YY42 pKa = 11.42VRR44 pKa = 11.84VCDD47 pKa = 3.88VYY49 pKa = 10.81GTGFYY54 pKa = 10.01YY55 pKa = 9.98IPGTEE60 pKa = 3.92TCLSINGYY68 pKa = 8.74VRR70 pKa = 11.84FQYY73 pKa = 10.71NVAGGIEE80 pKa = 4.43DD81 pKa = 5.31DD82 pKa = 4.58FGNDD86 pKa = 4.01LLTNSDD92 pKa = 3.37GDD94 pKa = 3.77NYY96 pKa = 10.59QAGSVVRR103 pKa = 11.84VRR105 pKa = 11.84LNFDD109 pKa = 2.97ARR111 pKa = 11.84EE112 pKa = 3.94EE113 pKa = 4.57TEE115 pKa = 4.58LGTLRR120 pKa = 11.84AYY122 pKa = 10.59ARR124 pKa = 11.84VQAQNAGGAYY134 pKa = 10.65SNDD137 pKa = 2.71ASYY140 pKa = 11.86NMDD143 pKa = 3.2MGFIQLGGLTMGYY156 pKa = 10.52LDD158 pKa = 4.17TLWTEE163 pKa = 4.38GDD165 pKa = 3.77GLLTDD170 pKa = 3.6TDD172 pKa = 4.77LPVGDD177 pKa = 4.24YY178 pKa = 7.2QTNRR182 pKa = 11.84ISYY185 pKa = 8.18TYY187 pKa = 10.27AANGFTAALSLEE199 pKa = 4.47DD200 pKa = 4.45DD201 pKa = 3.67ATGNFAPDD209 pKa = 3.32VVGRR213 pKa = 11.84LAYY216 pKa = 10.19AGGWGSVYY224 pKa = 10.65LAGSYY229 pKa = 10.52DD230 pKa = 3.68EE231 pKa = 4.99QVDD234 pKa = 3.96GRR236 pKa = 11.84LLTPASFTAAAASGLQVNSAGVIVPASATFVSGIDD271 pKa = 3.34PTTDD275 pKa = 3.08DD276 pKa = 4.15GAFALKK282 pKa = 10.53AGLSLTDD289 pKa = 5.44LIAADD294 pKa = 4.24SEE296 pKa = 4.54LKK298 pKa = 10.46IEE300 pKa = 4.32GSYY303 pKa = 11.35AFDD306 pKa = 4.1PSSYY310 pKa = 9.69STIGLFSNRR319 pKa = 11.84ATLGTQLDD327 pKa = 4.97LLTGTLPVEE336 pKa = 4.11WQIGAGYY343 pKa = 9.37AQQFGKK349 pKa = 10.79LGVAVSGVYY358 pKa = 10.82GEE360 pKa = 4.52TFDD363 pKa = 3.68TYY365 pKa = 10.95RR366 pKa = 11.84YY367 pKa = 8.03TNATTFSNRR376 pKa = 11.84GDD378 pKa = 3.37GEE380 pKa = 4.46YY381 pKa = 10.82YY382 pKa = 10.8KK383 pKa = 11.07LVGNVGYY390 pKa = 10.47EE391 pKa = 3.6ITNNFDD397 pKa = 3.11MLAEE401 pKa = 4.15ISYY404 pKa = 11.49ANIDD408 pKa = 3.46FDD410 pKa = 4.09GQNGFVDD417 pKa = 3.57NSLDD421 pKa = 3.41QTAGFLRR428 pKa = 11.84FVRR431 pKa = 11.84SFF433 pKa = 3.04
Molecular weight: 45.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1YI37|Q1YI37_AURMS Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase OS=Aurantimonas manganoxydans (strain ATCC BAA-1229 / DSM 21871 / SI85-9A1) OX=287752 GN=lpxA PE=3 SV=1
MM1 pKa = 6.75TAAIRR6 pKa = 11.84AAASWISWAVFPSSRR21 pKa = 11.84RR22 pKa = 11.84LPLAAPRR29 pKa = 11.84PRR31 pKa = 11.84RR32 pKa = 11.84HH33 pKa = 6.43RR34 pKa = 11.84SPPTHH39 pKa = 6.94RR40 pKa = 11.84ARR42 pKa = 11.84LRR44 pKa = 11.84RR45 pKa = 11.84CPTRR49 pKa = 11.84HH50 pKa = 5.7RR51 pKa = 11.84RR52 pKa = 11.84PRR54 pKa = 11.84PARR57 pKa = 11.84PPRR60 pKa = 11.84LRR62 pKa = 11.84RR63 pKa = 11.84SRR65 pKa = 11.84HH66 pKa = 4.15RR67 pKa = 11.84RR68 pKa = 11.84PRR70 pKa = 11.84KK71 pKa = 9.03FPHH74 pKa = 6.25RR75 pKa = 11.84PPRR78 pKa = 11.84QHH80 pKa = 7.44LSRR83 pKa = 11.84KK84 pKa = 8.6FLPTRR89 pKa = 11.84PRR91 pKa = 11.84MHH93 pKa = 7.06RR94 pKa = 11.84ASKK97 pKa = 8.29TRR99 pKa = 11.84RR100 pKa = 11.84PFRR103 pKa = 11.84PSRR106 pKa = 11.84SRR108 pKa = 11.84PQARR112 pKa = 11.84RR113 pKa = 11.84DD114 pKa = 3.58RR115 pKa = 11.84PTSRR119 pKa = 11.84PRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84TASSASFAHH132 pKa = 6.65
MM1 pKa = 6.75TAAIRR6 pKa = 11.84AAASWISWAVFPSSRR21 pKa = 11.84RR22 pKa = 11.84LPLAAPRR29 pKa = 11.84PRR31 pKa = 11.84RR32 pKa = 11.84HH33 pKa = 6.43RR34 pKa = 11.84SPPTHH39 pKa = 6.94RR40 pKa = 11.84ARR42 pKa = 11.84LRR44 pKa = 11.84RR45 pKa = 11.84CPTRR49 pKa = 11.84HH50 pKa = 5.7RR51 pKa = 11.84RR52 pKa = 11.84PRR54 pKa = 11.84PARR57 pKa = 11.84PPRR60 pKa = 11.84LRR62 pKa = 11.84RR63 pKa = 11.84SRR65 pKa = 11.84HH66 pKa = 4.15RR67 pKa = 11.84RR68 pKa = 11.84PRR70 pKa = 11.84KK71 pKa = 9.03FPHH74 pKa = 6.25RR75 pKa = 11.84PPRR78 pKa = 11.84QHH80 pKa = 7.44LSRR83 pKa = 11.84KK84 pKa = 8.6FLPTRR89 pKa = 11.84PRR91 pKa = 11.84MHH93 pKa = 7.06RR94 pKa = 11.84ASKK97 pKa = 8.29TRR99 pKa = 11.84RR100 pKa = 11.84PFRR103 pKa = 11.84PSRR106 pKa = 11.84SRR108 pKa = 11.84PQARR112 pKa = 11.84RR113 pKa = 11.84DD114 pKa = 3.58RR115 pKa = 11.84PTSRR119 pKa = 11.84PRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84TASSASFAHH132 pKa = 6.65
Molecular weight: 15.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1230513 |
62 |
3297 |
339.5 |
36.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.995 ± 0.056 | 0.86 ± 0.011 |
6.082 ± 0.036 | 5.842 ± 0.039 |
3.69 ± 0.025 | 8.941 ± 0.038 |
1.971 ± 0.021 | 5.125 ± 0.031 |
2.767 ± 0.034 | 9.671 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.54 ± 0.019 | 2.339 ± 0.022 |
5.22 ± 0.032 | 2.86 ± 0.025 |
7.622 ± 0.044 | 5.37 ± 0.034 |
5.393 ± 0.027 | 7.42 ± 0.029 |
1.207 ± 0.015 | 2.084 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
