
Pseudobutyrivibrio ruminis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Pseudobutyrivibrio
Average proteome isoelectric point is 5.62
Get precalculated fractions of proteins
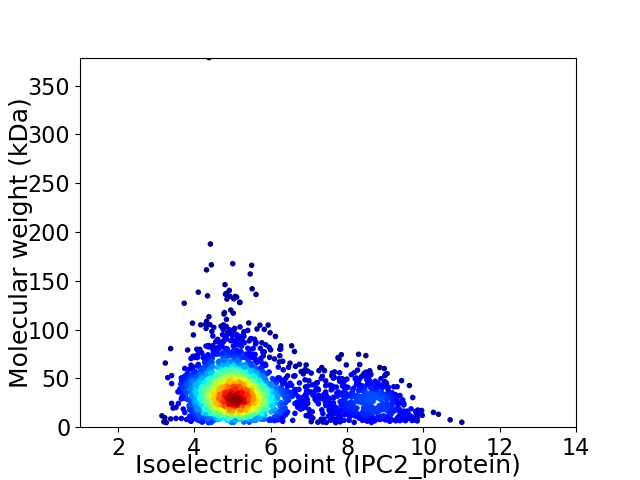
Virtual 2D-PAGE plot for 2521 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H7IIU3|A0A1H7IIU3_9FIRM Phosphatidate cytidylyltransferase OS=Pseudobutyrivibrio ruminis OX=46206 GN=SAMN02910377_01319 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.31KK3 pKa = 10.23KK4 pKa = 10.59LLALVLAATMVAGLAGCGNSSADD27 pKa = 3.55TASTGSASTASTSTDD42 pKa = 3.48LNVMIEE48 pKa = 4.26TAVEE52 pKa = 4.1SLDD55 pKa = 3.65PQQATDD61 pKa = 3.57GTSFEE66 pKa = 4.7VIADD70 pKa = 3.82YY71 pKa = 11.2TDD73 pKa = 4.31GLMQMDD79 pKa = 4.15SDD81 pKa = 4.32GNPINALAEE90 pKa = 4.38SYY92 pKa = 10.89DD93 pKa = 3.87LSDD96 pKa = 5.66DD97 pKa = 3.59GLTYY101 pKa = 10.06TFKK104 pKa = 10.71IRR106 pKa = 11.84DD107 pKa = 3.66DD108 pKa = 4.56ANWSNGEE115 pKa = 4.1PVTAQDD121 pKa = 6.31FVFAWQRR128 pKa = 11.84AVDD131 pKa = 3.83PEE133 pKa = 4.27VASEE137 pKa = 3.84YY138 pKa = 10.9SYY140 pKa = 10.83MLSDD144 pKa = 3.58IGQIVNAAEE153 pKa = 4.31IIAGEE158 pKa = 4.1KK159 pKa = 10.32DD160 pKa = 3.25KK161 pKa = 11.97SEE163 pKa = 4.61LGVTAVDD170 pKa = 4.05DD171 pKa = 3.81KK172 pKa = 11.34TLEE175 pKa = 4.07VQLNVPVSYY184 pKa = 10.12FLSLMYY190 pKa = 10.48FPTFYY195 pKa = 10.31PVNEE199 pKa = 4.19AFFNEE204 pKa = 4.57CGDD207 pKa = 3.88TFATSPEE214 pKa = 4.25TVLSNGAFVLDD225 pKa = 4.22SYY227 pKa = 11.36EE228 pKa = 3.99PAATAFHH235 pKa = 6.11LTKK238 pKa = 10.92NEE240 pKa = 4.06DD241 pKa = 3.72YY242 pKa = 11.04YY243 pKa = 11.62DD244 pKa = 3.98AASVSLSGLSYY255 pKa = 10.63QVIQDD260 pKa = 3.58SQTALMSYY268 pKa = 8.02QTGDD272 pKa = 3.47LDD274 pKa = 3.53ITLVNGDD281 pKa = 3.47QVDD284 pKa = 3.89QVEE287 pKa = 5.09DD288 pKa = 3.85DD289 pKa = 4.94PEE291 pKa = 4.23FQSIGAGYY299 pKa = 9.78LWYY302 pKa = 9.08VTPNIKK308 pKa = 9.8EE309 pKa = 4.16VPEE312 pKa = 3.93LANLNIRR319 pKa = 11.84LALTMAINRR328 pKa = 11.84EE329 pKa = 4.29SVTADD334 pKa = 3.27VLKK337 pKa = 10.96DD338 pKa = 3.61GSLPTYY344 pKa = 8.72TAIPRR349 pKa = 11.84DD350 pKa = 3.89FSADD354 pKa = 3.54FSADD358 pKa = 3.17QEE360 pKa = 4.37QFKK363 pKa = 10.66DD364 pKa = 3.6YY365 pKa = 11.18CRR367 pKa = 11.84DD368 pKa = 3.47DD369 pKa = 3.38AAAAAEE375 pKa = 4.13YY376 pKa = 8.58WQAGLDD382 pKa = 3.52EE383 pKa = 5.25LGITEE388 pKa = 4.41LTLEE392 pKa = 4.37LVADD396 pKa = 4.51EE397 pKa = 5.53DD398 pKa = 4.19DD399 pKa = 4.72APAKK403 pKa = 10.2VATVLKK409 pKa = 9.96EE410 pKa = 3.88EE411 pKa = 4.59WEE413 pKa = 4.37TTLPGLTVNITTEE426 pKa = 4.0PKK428 pKa = 9.2KK429 pKa = 10.85QRR431 pKa = 11.84VSDD434 pKa = 3.94LQEE437 pKa = 3.86GNYY440 pKa = 10.26EE441 pKa = 4.13LGLTRR446 pKa = 11.84WGPDD450 pKa = 2.85YY451 pKa = 10.75DD452 pKa = 5.83DD453 pKa = 4.26PMTYY457 pKa = 10.67LGMWYY462 pKa = 9.0TDD464 pKa = 3.13NPNNYY469 pKa = 9.04GFWSNAEE476 pKa = 3.55YY477 pKa = 10.71DD478 pKa = 4.43AILDD482 pKa = 3.75EE483 pKa = 4.86CTTGDD488 pKa = 3.72LCTEE492 pKa = 3.74IDD494 pKa = 3.37ARR496 pKa = 11.84RR497 pKa = 11.84DD498 pKa = 3.36RR499 pKa = 11.84LYY501 pKa = 10.9DD502 pKa = 3.41AEE504 pKa = 4.25QIVMEE509 pKa = 4.46EE510 pKa = 3.75AVIFPIYY517 pKa = 9.3TAANAQMVSSNVSGIDD533 pKa = 3.52FHH535 pKa = 7.18PVALNRR541 pKa = 11.84VYY543 pKa = 11.28KK544 pKa = 9.87NTTKK548 pKa = 10.93NN549 pKa = 3.07
MM1 pKa = 7.44KK2 pKa = 10.31KK3 pKa = 10.23KK4 pKa = 10.59LLALVLAATMVAGLAGCGNSSADD27 pKa = 3.55TASTGSASTASTSTDD42 pKa = 3.48LNVMIEE48 pKa = 4.26TAVEE52 pKa = 4.1SLDD55 pKa = 3.65PQQATDD61 pKa = 3.57GTSFEE66 pKa = 4.7VIADD70 pKa = 3.82YY71 pKa = 11.2TDD73 pKa = 4.31GLMQMDD79 pKa = 4.15SDD81 pKa = 4.32GNPINALAEE90 pKa = 4.38SYY92 pKa = 10.89DD93 pKa = 3.87LSDD96 pKa = 5.66DD97 pKa = 3.59GLTYY101 pKa = 10.06TFKK104 pKa = 10.71IRR106 pKa = 11.84DD107 pKa = 3.66DD108 pKa = 4.56ANWSNGEE115 pKa = 4.1PVTAQDD121 pKa = 6.31FVFAWQRR128 pKa = 11.84AVDD131 pKa = 3.83PEE133 pKa = 4.27VASEE137 pKa = 3.84YY138 pKa = 10.9SYY140 pKa = 10.83MLSDD144 pKa = 3.58IGQIVNAAEE153 pKa = 4.31IIAGEE158 pKa = 4.1KK159 pKa = 10.32DD160 pKa = 3.25KK161 pKa = 11.97SEE163 pKa = 4.61LGVTAVDD170 pKa = 4.05DD171 pKa = 3.81KK172 pKa = 11.34TLEE175 pKa = 4.07VQLNVPVSYY184 pKa = 10.12FLSLMYY190 pKa = 10.48FPTFYY195 pKa = 10.31PVNEE199 pKa = 4.19AFFNEE204 pKa = 4.57CGDD207 pKa = 3.88TFATSPEE214 pKa = 4.25TVLSNGAFVLDD225 pKa = 4.22SYY227 pKa = 11.36EE228 pKa = 3.99PAATAFHH235 pKa = 6.11LTKK238 pKa = 10.92NEE240 pKa = 4.06DD241 pKa = 3.72YY242 pKa = 11.04YY243 pKa = 11.62DD244 pKa = 3.98AASVSLSGLSYY255 pKa = 10.63QVIQDD260 pKa = 3.58SQTALMSYY268 pKa = 8.02QTGDD272 pKa = 3.47LDD274 pKa = 3.53ITLVNGDD281 pKa = 3.47QVDD284 pKa = 3.89QVEE287 pKa = 5.09DD288 pKa = 3.85DD289 pKa = 4.94PEE291 pKa = 4.23FQSIGAGYY299 pKa = 9.78LWYY302 pKa = 9.08VTPNIKK308 pKa = 9.8EE309 pKa = 4.16VPEE312 pKa = 3.93LANLNIRR319 pKa = 11.84LALTMAINRR328 pKa = 11.84EE329 pKa = 4.29SVTADD334 pKa = 3.27VLKK337 pKa = 10.96DD338 pKa = 3.61GSLPTYY344 pKa = 8.72TAIPRR349 pKa = 11.84DD350 pKa = 3.89FSADD354 pKa = 3.54FSADD358 pKa = 3.17QEE360 pKa = 4.37QFKK363 pKa = 10.66DD364 pKa = 3.6YY365 pKa = 11.18CRR367 pKa = 11.84DD368 pKa = 3.47DD369 pKa = 3.38AAAAAEE375 pKa = 4.13YY376 pKa = 8.58WQAGLDD382 pKa = 3.52EE383 pKa = 5.25LGITEE388 pKa = 4.41LTLEE392 pKa = 4.37LVADD396 pKa = 4.51EE397 pKa = 5.53DD398 pKa = 4.19DD399 pKa = 4.72APAKK403 pKa = 10.2VATVLKK409 pKa = 9.96EE410 pKa = 3.88EE411 pKa = 4.59WEE413 pKa = 4.37TTLPGLTVNITTEE426 pKa = 4.0PKK428 pKa = 9.2KK429 pKa = 10.85QRR431 pKa = 11.84VSDD434 pKa = 3.94LQEE437 pKa = 3.86GNYY440 pKa = 10.26EE441 pKa = 4.13LGLTRR446 pKa = 11.84WGPDD450 pKa = 2.85YY451 pKa = 10.75DD452 pKa = 5.83DD453 pKa = 4.26PMTYY457 pKa = 10.67LGMWYY462 pKa = 9.0TDD464 pKa = 3.13NPNNYY469 pKa = 9.04GFWSNAEE476 pKa = 3.55YY477 pKa = 10.71DD478 pKa = 4.43AILDD482 pKa = 3.75EE483 pKa = 4.86CTTGDD488 pKa = 3.72LCTEE492 pKa = 3.74IDD494 pKa = 3.37ARR496 pKa = 11.84RR497 pKa = 11.84DD498 pKa = 3.36RR499 pKa = 11.84LYY501 pKa = 10.9DD502 pKa = 3.41AEE504 pKa = 4.25QIVMEE509 pKa = 4.46EE510 pKa = 3.75AVIFPIYY517 pKa = 9.3TAANAQMVSSNVSGIDD533 pKa = 3.52FHH535 pKa = 7.18PVALNRR541 pKa = 11.84VYY543 pKa = 11.28KK544 pKa = 9.87NTTKK548 pKa = 10.93NN549 pKa = 3.07
Molecular weight: 60.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H7JNL3|A0A1H7JNL3_9FIRM Chromosome partitioning protein OS=Pseudobutyrivibrio ruminis OX=46206 GN=SAMN02910377_01749 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.0VHH16 pKa = 5.93GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.72VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84KK40 pKa = 8.85KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.0VHH16 pKa = 5.93GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.72VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84KK40 pKa = 8.85KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
841775 |
41 |
3439 |
333.9 |
37.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.603 ± 0.052 | 1.389 ± 0.018 |
6.427 ± 0.046 | 7.343 ± 0.046 |
4.249 ± 0.036 | 6.878 ± 0.046 |
1.612 ± 0.021 | 7.941 ± 0.048 |
6.742 ± 0.039 | 8.436 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.024 ± 0.027 | 4.93 ± 0.04 |
3.029 ± 0.029 | 2.811 ± 0.023 |
3.56 ± 0.036 | 6.126 ± 0.056 |
5.586 ± 0.055 | 7.135 ± 0.039 |
0.825 ± 0.017 | 4.354 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
