
Acetilactobacillus jinshanensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Acetilactobacillus
Average proteome isoelectric point is 8.42
Get precalculated fractions of proteins
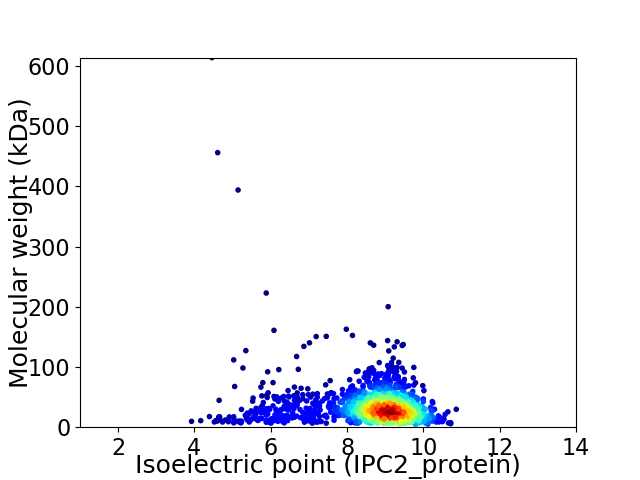
Virtual 2D-PAGE plot for 1450 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P6ZLK8|A0A4P6ZLK8_9LACO Glutamine synthetase OS=Acetilactobacillus jinshanensis OX=1720083 GN=glnA PE=3 SV=1
MM1 pKa = 7.71EE2 pKa = 5.44SLNSNQQISLTDD14 pKa = 3.35KK15 pKa = 11.03DD16 pKa = 4.19GNTVVYY22 pKa = 10.65NVLLTFKK29 pKa = 10.09STDD32 pKa = 3.7FHH34 pKa = 7.77KK35 pKa = 10.62SYY37 pKa = 11.11ILIYY41 pKa = 10.07PDD43 pKa = 3.87GRR45 pKa = 11.84PEE47 pKa = 3.83NQQVGVRR54 pKa = 11.84AFALPPDD61 pKa = 4.48EE62 pKa = 6.21DD63 pKa = 3.91PANPQSGDD71 pKa = 3.36LMPIEE76 pKa = 4.42TQKK79 pKa = 10.54EE80 pKa = 3.64WDD82 pKa = 3.58MVEE85 pKa = 4.4SVLNTFINPNEE96 pKa = 3.88
MM1 pKa = 7.71EE2 pKa = 5.44SLNSNQQISLTDD14 pKa = 3.35KK15 pKa = 11.03DD16 pKa = 4.19GNTVVYY22 pKa = 10.65NVLLTFKK29 pKa = 10.09STDD32 pKa = 3.7FHH34 pKa = 7.77KK35 pKa = 10.62SYY37 pKa = 11.11ILIYY41 pKa = 10.07PDD43 pKa = 3.87GRR45 pKa = 11.84PEE47 pKa = 3.83NQQVGVRR54 pKa = 11.84AFALPPDD61 pKa = 4.48EE62 pKa = 6.21DD63 pKa = 3.91PANPQSGDD71 pKa = 3.36LMPIEE76 pKa = 4.42TQKK79 pKa = 10.54EE80 pKa = 3.64WDD82 pKa = 3.58MVEE85 pKa = 4.4SVLNTFINPNEE96 pKa = 3.88
Molecular weight: 10.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P6ZJS9|A0A4P6ZJS9_9LACO Pyruvate kinase OS=Acetilactobacillus jinshanensis OX=1720083 GN=pyk PE=3 SV=1
MM1 pKa = 7.29VKK3 pKa = 10.27RR4 pKa = 11.84PVSNNIKK11 pKa = 10.59CPFCGTTFYY20 pKa = 10.88RR21 pKa = 11.84SDD23 pKa = 3.26LSYY26 pKa = 11.24VDD28 pKa = 3.0SRR30 pKa = 11.84VYY32 pKa = 10.29LKK34 pKa = 10.77RR35 pKa = 11.84MKK37 pKa = 10.39NRR39 pKa = 11.84LEE41 pKa = 3.98IYY43 pKa = 10.24PLGKK47 pKa = 10.0NVAASPKK54 pKa = 10.02LKK56 pKa = 9.75QQHH59 pKa = 6.58PEE61 pKa = 3.79LLRR64 pKa = 11.84LEE66 pKa = 4.52MSACPNCHH74 pKa = 6.95RR75 pKa = 11.84ITVRR79 pKa = 11.84AVGIGDD85 pKa = 3.63QYY87 pKa = 10.79KK88 pKa = 9.04HH89 pKa = 5.79HH90 pKa = 7.0HH91 pKa = 6.23VFNIYY96 pKa = 10.03PRR98 pKa = 11.84FNADD102 pKa = 3.51PLPTYY107 pKa = 10.83VPDD110 pKa = 5.48QIRR113 pKa = 11.84QDD115 pKa = 3.46YY116 pKa = 10.65KK117 pKa = 10.58EE118 pKa = 4.13ANEE121 pKa = 4.25VYY123 pKa = 9.93EE124 pKa = 4.05VSPNSAAMLARR135 pKa = 11.84RR136 pKa = 11.84ILEE139 pKa = 4.1EE140 pKa = 4.79IIADD144 pKa = 4.73FYY146 pKa = 11.26HH147 pKa = 8.03IEE149 pKa = 4.19EE150 pKa = 4.86GDD152 pKa = 3.87LYY154 pKa = 11.25HH155 pKa = 7.78DD156 pKa = 5.41LDD158 pKa = 3.97QLRR161 pKa = 11.84HH162 pKa = 5.39KK163 pKa = 10.57QSNPRR168 pKa = 11.84VWQAIDD174 pKa = 3.43SIRR177 pKa = 11.84RR178 pKa = 11.84LGNIGAHH185 pKa = 4.36QTQNVDD191 pKa = 3.18NVFGNVSVKK200 pKa = 9.79GAKK203 pKa = 9.77EE204 pKa = 3.73IIGLIRR210 pKa = 11.84MVIHH214 pKa = 6.3DD215 pKa = 4.7TYY217 pKa = 11.77VEE219 pKa = 4.16DD220 pKa = 3.42HH221 pKa = 6.99LSRR224 pKa = 11.84HH225 pKa = 5.49LEE227 pKa = 3.81KK228 pKa = 10.94SVIRR232 pKa = 11.84SAKK235 pKa = 8.54KK236 pKa = 10.33LNYY239 pKa = 8.67QPHH242 pKa = 6.82YY243 pKa = 9.9YY244 pKa = 9.93PSDD247 pKa = 3.66EE248 pKa = 4.3NSQSSRR254 pKa = 11.84RR255 pKa = 11.84NYY257 pKa = 10.17RR258 pKa = 11.84SRR260 pKa = 11.84NTHH263 pKa = 5.84KK264 pKa = 10.61SYY266 pKa = 11.24SSHH269 pKa = 6.25NSNNANRR276 pKa = 11.84SKK278 pKa = 10.89SSNASRR284 pKa = 11.84GHH286 pKa = 4.22QQHH289 pKa = 6.66RR290 pKa = 11.84NNHH293 pKa = 5.31RR294 pKa = 11.84RR295 pKa = 11.84NQNNSRR301 pKa = 11.84RR302 pKa = 11.84EE303 pKa = 3.78NNRR306 pKa = 11.84RR307 pKa = 11.84SNQKK311 pKa = 10.1RR312 pKa = 11.84NIRR315 pKa = 11.84RR316 pKa = 11.84NSSTPKK322 pKa = 9.02RR323 pKa = 11.84NNYY326 pKa = 9.07RR327 pKa = 11.84KK328 pKa = 9.57QSRR331 pKa = 11.84PSNNRR336 pKa = 11.84KK337 pKa = 9.61SNQRR341 pKa = 11.84NSNSYY346 pKa = 10.75RR347 pKa = 11.84KK348 pKa = 6.69THH350 pKa = 6.56RR351 pKa = 11.84NNNYY355 pKa = 9.21RR356 pKa = 11.84RR357 pKa = 11.84STRR360 pKa = 11.84SRR362 pKa = 11.84EE363 pKa = 3.78RR364 pKa = 11.84NQQSRR369 pKa = 11.84SRR371 pKa = 11.84HH372 pKa = 3.77TQRR375 pKa = 11.84RR376 pKa = 11.84STRR379 pKa = 11.84HH380 pKa = 4.62SKK382 pKa = 10.32KK383 pKa = 10.29SRR385 pKa = 11.84VRR387 pKa = 11.84DD388 pKa = 3.4KK389 pKa = 11.26SFTIIQGKK397 pKa = 9.02
MM1 pKa = 7.29VKK3 pKa = 10.27RR4 pKa = 11.84PVSNNIKK11 pKa = 10.59CPFCGTTFYY20 pKa = 10.88RR21 pKa = 11.84SDD23 pKa = 3.26LSYY26 pKa = 11.24VDD28 pKa = 3.0SRR30 pKa = 11.84VYY32 pKa = 10.29LKK34 pKa = 10.77RR35 pKa = 11.84MKK37 pKa = 10.39NRR39 pKa = 11.84LEE41 pKa = 3.98IYY43 pKa = 10.24PLGKK47 pKa = 10.0NVAASPKK54 pKa = 10.02LKK56 pKa = 9.75QQHH59 pKa = 6.58PEE61 pKa = 3.79LLRR64 pKa = 11.84LEE66 pKa = 4.52MSACPNCHH74 pKa = 6.95RR75 pKa = 11.84ITVRR79 pKa = 11.84AVGIGDD85 pKa = 3.63QYY87 pKa = 10.79KK88 pKa = 9.04HH89 pKa = 5.79HH90 pKa = 7.0HH91 pKa = 6.23VFNIYY96 pKa = 10.03PRR98 pKa = 11.84FNADD102 pKa = 3.51PLPTYY107 pKa = 10.83VPDD110 pKa = 5.48QIRR113 pKa = 11.84QDD115 pKa = 3.46YY116 pKa = 10.65KK117 pKa = 10.58EE118 pKa = 4.13ANEE121 pKa = 4.25VYY123 pKa = 9.93EE124 pKa = 4.05VSPNSAAMLARR135 pKa = 11.84RR136 pKa = 11.84ILEE139 pKa = 4.1EE140 pKa = 4.79IIADD144 pKa = 4.73FYY146 pKa = 11.26HH147 pKa = 8.03IEE149 pKa = 4.19EE150 pKa = 4.86GDD152 pKa = 3.87LYY154 pKa = 11.25HH155 pKa = 7.78DD156 pKa = 5.41LDD158 pKa = 3.97QLRR161 pKa = 11.84HH162 pKa = 5.39KK163 pKa = 10.57QSNPRR168 pKa = 11.84VWQAIDD174 pKa = 3.43SIRR177 pKa = 11.84RR178 pKa = 11.84LGNIGAHH185 pKa = 4.36QTQNVDD191 pKa = 3.18NVFGNVSVKK200 pKa = 9.79GAKK203 pKa = 9.77EE204 pKa = 3.73IIGLIRR210 pKa = 11.84MVIHH214 pKa = 6.3DD215 pKa = 4.7TYY217 pKa = 11.77VEE219 pKa = 4.16DD220 pKa = 3.42HH221 pKa = 6.99LSRR224 pKa = 11.84HH225 pKa = 5.49LEE227 pKa = 3.81KK228 pKa = 10.94SVIRR232 pKa = 11.84SAKK235 pKa = 8.54KK236 pKa = 10.33LNYY239 pKa = 8.67QPHH242 pKa = 6.82YY243 pKa = 9.9YY244 pKa = 9.93PSDD247 pKa = 3.66EE248 pKa = 4.3NSQSSRR254 pKa = 11.84RR255 pKa = 11.84NYY257 pKa = 10.17RR258 pKa = 11.84SRR260 pKa = 11.84NTHH263 pKa = 5.84KK264 pKa = 10.61SYY266 pKa = 11.24SSHH269 pKa = 6.25NSNNANRR276 pKa = 11.84SKK278 pKa = 10.89SSNASRR284 pKa = 11.84GHH286 pKa = 4.22QQHH289 pKa = 6.66RR290 pKa = 11.84NNHH293 pKa = 5.31RR294 pKa = 11.84RR295 pKa = 11.84NQNNSRR301 pKa = 11.84RR302 pKa = 11.84EE303 pKa = 3.78NNRR306 pKa = 11.84RR307 pKa = 11.84SNQKK311 pKa = 10.1RR312 pKa = 11.84NIRR315 pKa = 11.84RR316 pKa = 11.84NSSTPKK322 pKa = 9.02RR323 pKa = 11.84NNYY326 pKa = 9.07RR327 pKa = 11.84KK328 pKa = 9.57QSRR331 pKa = 11.84PSNNRR336 pKa = 11.84KK337 pKa = 9.61SNQRR341 pKa = 11.84NSNSYY346 pKa = 10.75RR347 pKa = 11.84KK348 pKa = 6.69THH350 pKa = 6.56RR351 pKa = 11.84NNNYY355 pKa = 9.21RR356 pKa = 11.84RR357 pKa = 11.84STRR360 pKa = 11.84SRR362 pKa = 11.84EE363 pKa = 3.78RR364 pKa = 11.84NQQSRR369 pKa = 11.84SRR371 pKa = 11.84HH372 pKa = 3.77TQRR375 pKa = 11.84RR376 pKa = 11.84STRR379 pKa = 11.84HH380 pKa = 4.62SKK382 pKa = 10.32KK383 pKa = 10.29SRR385 pKa = 11.84VRR387 pKa = 11.84DD388 pKa = 3.4KK389 pKa = 11.26SFTIIQGKK397 pKa = 9.02
Molecular weight: 46.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
454499 |
39 |
6060 |
313.4 |
35.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.043 ± 0.159 | 0.676 ± 0.02 |
5.296 ± 0.06 | 3.411 ± 0.043 |
4.105 ± 0.062 | 6.306 ± 0.059 |
3.34 ± 0.043 | 7.507 ± 0.089 |
8.072 ± 0.099 | 8.995 ± 0.107 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.859 ± 0.045 | 5.988 ± 0.094 |
3.901 ± 0.048 | 4.162 ± 0.051 |
4.313 ± 0.079 | 6.681 ± 0.233 |
5.45 ± 0.06 | 6.774 ± 0.056 |
1.034 ± 0.021 | 4.086 ± 0.048 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
