
Escherichia phage Pollock
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Schitoviridae; Humphriesvirinae; Pollockvirus; Escherichia virus Pollock
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
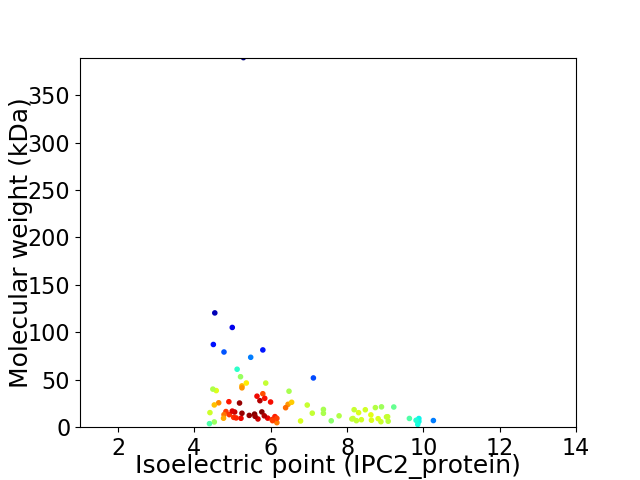
Virtual 2D-PAGE plot for 84 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0YR07|A0A0A0YR07_9CAUD Uncharacterized protein OS=Escherichia phage Pollock OX=1540097 GN=CPT_Pollock67 PE=4 SV=1
MM1 pKa = 7.48EE2 pKa = 5.28LVLHH6 pKa = 6.19IKK8 pKa = 10.01EE9 pKa = 4.43RR10 pKa = 11.84GRR12 pKa = 11.84KK13 pKa = 7.84PPLMYY18 pKa = 9.96MGLFSRR24 pKa = 11.84KK25 pKa = 9.47KK26 pKa = 8.44IISVSSVTVNLAGDD40 pKa = 3.6DD41 pKa = 4.32AEE43 pKa = 5.28NFLQKK48 pKa = 9.79TITTAVMNGSGIGSTLNQAYY68 pKa = 10.0LNGMGTQIRR77 pKa = 11.84QAYY80 pKa = 9.49RR81 pKa = 11.84YY82 pKa = 9.47GRR84 pKa = 11.84DD85 pKa = 3.62YY86 pKa = 11.62YY87 pKa = 11.65SLGLPDD93 pKa = 4.7GSIFYY98 pKa = 9.2VVPNYY103 pKa = 10.6DD104 pKa = 3.7DD105 pKa = 4.97LMTVLSSLNEE115 pKa = 3.98GKK117 pKa = 10.05EE118 pKa = 4.04VSIVMSDD125 pKa = 3.65YY126 pKa = 11.35NVADD130 pKa = 4.08LSYY133 pKa = 9.87WIEE136 pKa = 3.98RR137 pKa = 11.84YY138 pKa = 7.93LTNTYY143 pKa = 10.81DD144 pKa = 2.91WDD146 pKa = 3.73ADD148 pKa = 3.77YY149 pKa = 11.56GGMGNPPAGVASTATIDD166 pKa = 3.62WSIDD170 pKa = 3.2NNGVVTITMGTGSEE184 pKa = 3.81IDD186 pKa = 3.48FTEE189 pKa = 3.94QVSFSDD195 pKa = 3.15INYY198 pKa = 9.05GSEE201 pKa = 4.51YY202 pKa = 8.9YY203 pKa = 9.85QVIYY207 pKa = 10.29RR208 pKa = 11.84VRR210 pKa = 11.84TPGTPEE216 pKa = 3.55VTTEE220 pKa = 3.52TRR222 pKa = 11.84AYY224 pKa = 10.37EE225 pKa = 4.27EE226 pKa = 4.52GDD228 pKa = 3.65EE229 pKa = 4.44EE230 pKa = 5.03GTTTDD235 pKa = 3.25EE236 pKa = 4.57TINNNFGLITSTVTTVQATINEE258 pKa = 4.64DD259 pKa = 3.25KK260 pKa = 11.02TEE262 pKa = 4.16TTITTTVVVSTLSRR276 pKa = 11.84KK277 pKa = 9.82KK278 pKa = 10.36YY279 pKa = 10.1FIYY282 pKa = 10.29EE283 pKa = 4.41AGSGTYY289 pKa = 9.96PILDD293 pKa = 4.44KK294 pKa = 11.03ILTDD298 pKa = 3.48VVRR301 pKa = 11.84DD302 pKa = 3.46SAYY305 pKa = 10.57YY306 pKa = 8.85PVVPLRR312 pKa = 11.84VNNTDD317 pKa = 3.07LTDD320 pKa = 4.35PNNVSAEE327 pKa = 3.94QFKK330 pKa = 10.2TSKK333 pKa = 9.71TLLNKK338 pKa = 10.35LNLRR342 pKa = 11.84FTDD345 pKa = 5.18LGDD348 pKa = 3.91TLNEE352 pKa = 4.01NPDD355 pKa = 3.4IKK357 pKa = 11.1DD358 pKa = 2.9IDD360 pKa = 3.61HH361 pKa = 7.06AFFVMGISLNSKK373 pKa = 8.71YY374 pKa = 10.22EE375 pKa = 3.74SSIDD379 pKa = 3.56YY380 pKa = 10.23LHH382 pKa = 7.09EE383 pKa = 4.24FFKK386 pKa = 11.4YY387 pKa = 10.05LAQVSPNDD395 pKa = 3.39KK396 pKa = 10.2EE397 pKa = 5.76NYY399 pKa = 8.2IKK401 pKa = 10.37WYY403 pKa = 8.04TEE405 pKa = 3.64NVNEE409 pKa = 4.82EE410 pKa = 4.21GTISNTDD417 pKa = 2.88SKK419 pKa = 11.3PPVNRR424 pKa = 11.84LSLRR428 pKa = 11.84QDD430 pKa = 3.45PYY432 pKa = 11.3DD433 pKa = 3.21ITIAYY438 pKa = 8.86QYY440 pKa = 11.63SDD442 pKa = 3.63LTVKK446 pKa = 10.61NGSIGSIGTVTRR458 pKa = 11.84SVGNKK463 pKa = 8.18ATINVVNILNASVEE477 pKa = 4.2MTADD481 pKa = 2.86VSTILFRR488 pKa = 11.84KK489 pKa = 9.41QISEE493 pKa = 4.09TQYY496 pKa = 11.26EE497 pKa = 4.19EE498 pKa = 5.07VEE500 pKa = 4.19ICGLEE505 pKa = 4.22HH506 pKa = 6.89INYY509 pKa = 9.0IYY511 pKa = 10.36KK512 pKa = 10.12GHH514 pKa = 6.16SVNITGEE521 pKa = 4.07ASLSDD526 pKa = 3.83PDD528 pKa = 4.22NEE530 pKa = 4.41GFLIPLCANIVDD542 pKa = 4.04AQQIVHH548 pKa = 6.29RR549 pKa = 11.84TQMTYY554 pKa = 10.7DD555 pKa = 3.66CLHH558 pKa = 6.5IVVNSYY564 pKa = 10.26QVTKK568 pKa = 10.83AKK570 pKa = 9.69WYY572 pKa = 8.81QSGIFKK578 pKa = 10.7VVTIAIAVVIAVYY591 pKa = 10.87SLGTLSAGITAAASAATAAGTSVAAAVAVYY621 pKa = 8.68ITTQVAIGLAISFGVSILAKK641 pKa = 9.77FVSPNIMFIATIAMLTYY658 pKa = 10.16GAVSAYY664 pKa = 9.87QSYY667 pKa = 8.19GTTGNKK673 pKa = 9.07GLPYY677 pKa = 9.45ATEE680 pKa = 4.01VMNLVPSVAKK690 pKa = 9.81GAQMRR695 pKa = 11.84LQQDD699 pKa = 3.52LQNIVKK705 pKa = 9.88EE706 pKa = 4.12MQKK709 pKa = 10.25NAEE712 pKa = 4.08NYY714 pKa = 7.8KK715 pKa = 10.47KK716 pKa = 10.41QMEE719 pKa = 4.64EE720 pKa = 4.14IEE722 pKa = 4.44DD723 pKa = 3.69AMDD726 pKa = 4.23ALGTINPNIDD736 pKa = 2.87IDD738 pKa = 4.05ALINSAFFNLFEE750 pKa = 4.42QPDD753 pKa = 3.67EE754 pKa = 4.4FFTRR758 pKa = 11.84TLSTNPGVVTLEE770 pKa = 4.28SIEE773 pKa = 4.61NYY775 pKa = 9.98VDD777 pKa = 3.54YY778 pKa = 11.19ALTLPTDD785 pKa = 4.35LNSMRR790 pKa = 11.84SS791 pKa = 3.34
MM1 pKa = 7.48EE2 pKa = 5.28LVLHH6 pKa = 6.19IKK8 pKa = 10.01EE9 pKa = 4.43RR10 pKa = 11.84GRR12 pKa = 11.84KK13 pKa = 7.84PPLMYY18 pKa = 9.96MGLFSRR24 pKa = 11.84KK25 pKa = 9.47KK26 pKa = 8.44IISVSSVTVNLAGDD40 pKa = 3.6DD41 pKa = 4.32AEE43 pKa = 5.28NFLQKK48 pKa = 9.79TITTAVMNGSGIGSTLNQAYY68 pKa = 10.0LNGMGTQIRR77 pKa = 11.84QAYY80 pKa = 9.49RR81 pKa = 11.84YY82 pKa = 9.47GRR84 pKa = 11.84DD85 pKa = 3.62YY86 pKa = 11.62YY87 pKa = 11.65SLGLPDD93 pKa = 4.7GSIFYY98 pKa = 9.2VVPNYY103 pKa = 10.6DD104 pKa = 3.7DD105 pKa = 4.97LMTVLSSLNEE115 pKa = 3.98GKK117 pKa = 10.05EE118 pKa = 4.04VSIVMSDD125 pKa = 3.65YY126 pKa = 11.35NVADD130 pKa = 4.08LSYY133 pKa = 9.87WIEE136 pKa = 3.98RR137 pKa = 11.84YY138 pKa = 7.93LTNTYY143 pKa = 10.81DD144 pKa = 2.91WDD146 pKa = 3.73ADD148 pKa = 3.77YY149 pKa = 11.56GGMGNPPAGVASTATIDD166 pKa = 3.62WSIDD170 pKa = 3.2NNGVVTITMGTGSEE184 pKa = 3.81IDD186 pKa = 3.48FTEE189 pKa = 3.94QVSFSDD195 pKa = 3.15INYY198 pKa = 9.05GSEE201 pKa = 4.51YY202 pKa = 8.9YY203 pKa = 9.85QVIYY207 pKa = 10.29RR208 pKa = 11.84VRR210 pKa = 11.84TPGTPEE216 pKa = 3.55VTTEE220 pKa = 3.52TRR222 pKa = 11.84AYY224 pKa = 10.37EE225 pKa = 4.27EE226 pKa = 4.52GDD228 pKa = 3.65EE229 pKa = 4.44EE230 pKa = 5.03GTTTDD235 pKa = 3.25EE236 pKa = 4.57TINNNFGLITSTVTTVQATINEE258 pKa = 4.64DD259 pKa = 3.25KK260 pKa = 11.02TEE262 pKa = 4.16TTITTTVVVSTLSRR276 pKa = 11.84KK277 pKa = 9.82KK278 pKa = 10.36YY279 pKa = 10.1FIYY282 pKa = 10.29EE283 pKa = 4.41AGSGTYY289 pKa = 9.96PILDD293 pKa = 4.44KK294 pKa = 11.03ILTDD298 pKa = 3.48VVRR301 pKa = 11.84DD302 pKa = 3.46SAYY305 pKa = 10.57YY306 pKa = 8.85PVVPLRR312 pKa = 11.84VNNTDD317 pKa = 3.07LTDD320 pKa = 4.35PNNVSAEE327 pKa = 3.94QFKK330 pKa = 10.2TSKK333 pKa = 9.71TLLNKK338 pKa = 10.35LNLRR342 pKa = 11.84FTDD345 pKa = 5.18LGDD348 pKa = 3.91TLNEE352 pKa = 4.01NPDD355 pKa = 3.4IKK357 pKa = 11.1DD358 pKa = 2.9IDD360 pKa = 3.61HH361 pKa = 7.06AFFVMGISLNSKK373 pKa = 8.71YY374 pKa = 10.22EE375 pKa = 3.74SSIDD379 pKa = 3.56YY380 pKa = 10.23LHH382 pKa = 7.09EE383 pKa = 4.24FFKK386 pKa = 11.4YY387 pKa = 10.05LAQVSPNDD395 pKa = 3.39KK396 pKa = 10.2EE397 pKa = 5.76NYY399 pKa = 8.2IKK401 pKa = 10.37WYY403 pKa = 8.04TEE405 pKa = 3.64NVNEE409 pKa = 4.82EE410 pKa = 4.21GTISNTDD417 pKa = 2.88SKK419 pKa = 11.3PPVNRR424 pKa = 11.84LSLRR428 pKa = 11.84QDD430 pKa = 3.45PYY432 pKa = 11.3DD433 pKa = 3.21ITIAYY438 pKa = 8.86QYY440 pKa = 11.63SDD442 pKa = 3.63LTVKK446 pKa = 10.61NGSIGSIGTVTRR458 pKa = 11.84SVGNKK463 pKa = 8.18ATINVVNILNASVEE477 pKa = 4.2MTADD481 pKa = 2.86VSTILFRR488 pKa = 11.84KK489 pKa = 9.41QISEE493 pKa = 4.09TQYY496 pKa = 11.26EE497 pKa = 4.19EE498 pKa = 5.07VEE500 pKa = 4.19ICGLEE505 pKa = 4.22HH506 pKa = 6.89INYY509 pKa = 9.0IYY511 pKa = 10.36KK512 pKa = 10.12GHH514 pKa = 6.16SVNITGEE521 pKa = 4.07ASLSDD526 pKa = 3.83PDD528 pKa = 4.22NEE530 pKa = 4.41GFLIPLCANIVDD542 pKa = 4.04AQQIVHH548 pKa = 6.29RR549 pKa = 11.84TQMTYY554 pKa = 10.7DD555 pKa = 3.66CLHH558 pKa = 6.5IVVNSYY564 pKa = 10.26QVTKK568 pKa = 10.83AKK570 pKa = 9.69WYY572 pKa = 8.81QSGIFKK578 pKa = 10.7VVTIAIAVVIAVYY591 pKa = 10.87SLGTLSAGITAAASAATAAGTSVAAAVAVYY621 pKa = 8.68ITTQVAIGLAISFGVSILAKK641 pKa = 9.77FVSPNIMFIATIAMLTYY658 pKa = 10.16GAVSAYY664 pKa = 9.87QSYY667 pKa = 8.19GTTGNKK673 pKa = 9.07GLPYY677 pKa = 9.45ATEE680 pKa = 4.01VMNLVPSVAKK690 pKa = 9.81GAQMRR695 pKa = 11.84LQQDD699 pKa = 3.52LQNIVKK705 pKa = 9.88EE706 pKa = 4.12MQKK709 pKa = 10.25NAEE712 pKa = 4.08NYY714 pKa = 7.8KK715 pKa = 10.47KK716 pKa = 10.41QMEE719 pKa = 4.64EE720 pKa = 4.14IEE722 pKa = 4.44DD723 pKa = 3.69AMDD726 pKa = 4.23ALGTINPNIDD736 pKa = 2.87IDD738 pKa = 4.05ALINSAFFNLFEE750 pKa = 4.42QPDD753 pKa = 3.67EE754 pKa = 4.4FFTRR758 pKa = 11.84TLSTNPGVVTLEE770 pKa = 4.28SIEE773 pKa = 4.61NYY775 pKa = 9.98VDD777 pKa = 3.54YY778 pKa = 11.19ALTLPTDD785 pKa = 4.35LNSMRR790 pKa = 11.84SS791 pKa = 3.34
Molecular weight: 87.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0YUA7|A0A0A0YUA7_9CAUD Uncharacterized protein OS=Escherichia phage Pollock OX=1540097 GN=CPT_Pollock79 PE=4 SV=1
MM1 pKa = 7.71AKK3 pKa = 8.95LTSKK7 pKa = 10.11QRR9 pKa = 11.84NKK11 pKa = 10.82LPDD14 pKa = 3.33SAFAGPDD21 pKa = 2.66RR22 pKa = 11.84SYY24 pKa = 10.94PVHH27 pKa = 7.29DD28 pKa = 4.44KK29 pKa = 11.39AHH31 pKa = 6.41AANAKK36 pKa = 9.8ARR38 pKa = 11.84ATQQYY43 pKa = 9.28EE44 pKa = 3.89AGNLSKK50 pKa = 10.69SQRR53 pKa = 11.84DD54 pKa = 3.89KK55 pKa = 11.08IRR57 pKa = 11.84ARR59 pKa = 11.84ANKK62 pKa = 9.37VLYY65 pKa = 10.4GG66 pKa = 3.64
MM1 pKa = 7.71AKK3 pKa = 8.95LTSKK7 pKa = 10.11QRR9 pKa = 11.84NKK11 pKa = 10.82LPDD14 pKa = 3.33SAFAGPDD21 pKa = 2.66RR22 pKa = 11.84SYY24 pKa = 10.94PVHH27 pKa = 7.29DD28 pKa = 4.44KK29 pKa = 11.39AHH31 pKa = 6.41AANAKK36 pKa = 9.8ARR38 pKa = 11.84ATQQYY43 pKa = 9.28EE44 pKa = 3.89AGNLSKK50 pKa = 10.69SQRR53 pKa = 11.84DD54 pKa = 3.89KK55 pKa = 11.08IRR57 pKa = 11.84ARR59 pKa = 11.84ANKK62 pKa = 9.37VLYY65 pKa = 10.4GG66 pKa = 3.64
Molecular weight: 7.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
20944 |
28 |
3537 |
249.3 |
27.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.477 ± 0.461 | 1.003 ± 0.185 |
5.567 ± 0.198 | 7.066 ± 0.239 |
3.71 ± 0.167 | 5.954 ± 0.233 |
1.681 ± 0.141 | 7.176 ± 0.239 |
6.699 ± 0.32 | 8.79 ± 0.232 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.617 ± 0.151 | 5.949 ± 0.224 |
3.834 ± 0.135 | 4.311 ± 0.352 |
4.206 ± 0.183 | 6.431 ± 0.175 |
6.255 ± 0.278 | 6.403 ± 0.2 |
1.074 ± 0.124 | 3.796 ± 0.261 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
