
Myotis polyomavirus VM-2008
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus; Myotis lucifugus polyomavirus 1
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
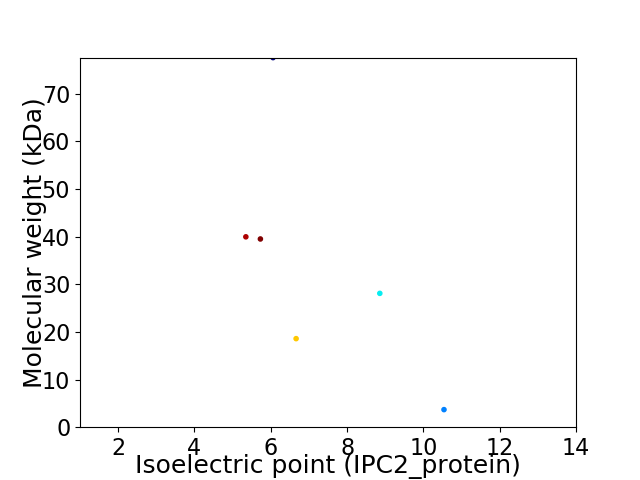
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B6E000|B6E000_9POLY Minor capsid protein OS=Myotis polyomavirus VM-2008 OX=563775 PE=3 SV=1
MM1 pKa = 7.38GALLTVLAEE10 pKa = 4.4VIEE13 pKa = 4.3LASVTGLSAEE23 pKa = 4.43SIISGEE29 pKa = 4.03AFATAEE35 pKa = 4.25LLEE38 pKa = 4.17SHH40 pKa = 7.11IANLVTYY47 pKa = 10.3GGLTEE52 pKa = 4.96AEE54 pKa = 3.92ALAATEE60 pKa = 4.16VTTEE64 pKa = 4.03AFEE67 pKa = 4.51ALQSLTPNFSQVFGAIAGIDD87 pKa = 3.15IAANSSLFLGAAVAAALYY105 pKa = 8.64PYY107 pKa = 10.13NWDD110 pKa = 3.32YY111 pKa = 11.26SQPIAMANMDD121 pKa = 4.14YY122 pKa = 10.56PIVLYY127 pKa = 10.78HH128 pKa = 7.39PDD130 pKa = 3.63FDD132 pKa = 4.65IDD134 pKa = 4.24FPGLRR139 pKa = 11.84TLARR143 pKa = 11.84FIHH146 pKa = 6.33YY147 pKa = 9.88ISPEE151 pKa = 3.49HH152 pKa = 6.44WGADD156 pKa = 3.4LFRR159 pKa = 11.84AMGNYY164 pKa = 9.99LYY166 pKa = 10.8EE167 pKa = 4.09LLQRR171 pKa = 11.84EE172 pKa = 4.43RR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84QIDD178 pKa = 3.07RR179 pKa = 11.84MTRR182 pKa = 11.84EE183 pKa = 3.63LARR186 pKa = 11.84RR187 pKa = 11.84SGEE190 pKa = 3.98EE191 pKa = 3.64VADD194 pKa = 3.98RR195 pKa = 11.84LARR198 pKa = 11.84YY199 pKa = 9.22FEE201 pKa = 3.92NARR204 pKa = 11.84WAVTNAISTPVTIYY218 pKa = 10.5RR219 pKa = 11.84YY220 pKa = 10.37LGDD223 pKa = 3.85YY224 pKa = 10.18YY225 pKa = 10.92RR226 pKa = 11.84ALPGPNPIRR235 pKa = 11.84ARR237 pKa = 11.84QLARR241 pKa = 11.84SLGQEE246 pKa = 3.46EE247 pKa = 5.4PYY249 pKa = 10.39RR250 pKa = 11.84YY251 pKa = 10.31DD252 pKa = 3.46RR253 pKa = 11.84FEE255 pKa = 4.33TEE257 pKa = 3.45HH258 pKa = 6.84GEE260 pKa = 3.95PSFTPKK266 pKa = 10.06QKK268 pKa = 10.36SADD271 pKa = 3.5YY272 pKa = 10.32VEE274 pKa = 5.85RR275 pKa = 11.84YY276 pKa = 7.48EE277 pKa = 4.4PPGGAYY283 pKa = 9.39QRR285 pKa = 11.84NTPDD289 pKa = 2.53WMLPLILGLYY299 pKa = 10.37GDD301 pKa = 5.24ISPSWGDD308 pKa = 3.14TIEE311 pKa = 4.1RR312 pKa = 11.84VEE314 pKa = 4.13EE315 pKa = 4.13EE316 pKa = 4.32EE317 pKa = 5.02KK318 pKa = 11.11EE319 pKa = 4.24EE320 pKa = 4.15EE321 pKa = 4.85DD322 pKa = 4.14GPKK325 pKa = 9.92KK326 pKa = 10.44KK327 pKa = 10.28KK328 pKa = 10.2LRR330 pKa = 11.84TVRR333 pKa = 11.84GPKK336 pKa = 8.95TNNKK340 pKa = 8.73RR341 pKa = 11.84RR342 pKa = 11.84NRR344 pKa = 11.84STQRR348 pKa = 11.84PNRR351 pKa = 11.84PRR353 pKa = 4.21
MM1 pKa = 7.38GALLTVLAEE10 pKa = 4.4VIEE13 pKa = 4.3LASVTGLSAEE23 pKa = 4.43SIISGEE29 pKa = 4.03AFATAEE35 pKa = 4.25LLEE38 pKa = 4.17SHH40 pKa = 7.11IANLVTYY47 pKa = 10.3GGLTEE52 pKa = 4.96AEE54 pKa = 3.92ALAATEE60 pKa = 4.16VTTEE64 pKa = 4.03AFEE67 pKa = 4.51ALQSLTPNFSQVFGAIAGIDD87 pKa = 3.15IAANSSLFLGAAVAAALYY105 pKa = 8.64PYY107 pKa = 10.13NWDD110 pKa = 3.32YY111 pKa = 11.26SQPIAMANMDD121 pKa = 4.14YY122 pKa = 10.56PIVLYY127 pKa = 10.78HH128 pKa = 7.39PDD130 pKa = 3.63FDD132 pKa = 4.65IDD134 pKa = 4.24FPGLRR139 pKa = 11.84TLARR143 pKa = 11.84FIHH146 pKa = 6.33YY147 pKa = 9.88ISPEE151 pKa = 3.49HH152 pKa = 6.44WGADD156 pKa = 3.4LFRR159 pKa = 11.84AMGNYY164 pKa = 9.99LYY166 pKa = 10.8EE167 pKa = 4.09LLQRR171 pKa = 11.84EE172 pKa = 4.43RR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84QIDD178 pKa = 3.07RR179 pKa = 11.84MTRR182 pKa = 11.84EE183 pKa = 3.63LARR186 pKa = 11.84RR187 pKa = 11.84SGEE190 pKa = 3.98EE191 pKa = 3.64VADD194 pKa = 3.98RR195 pKa = 11.84LARR198 pKa = 11.84YY199 pKa = 9.22FEE201 pKa = 3.92NARR204 pKa = 11.84WAVTNAISTPVTIYY218 pKa = 10.5RR219 pKa = 11.84YY220 pKa = 10.37LGDD223 pKa = 3.85YY224 pKa = 10.18YY225 pKa = 10.92RR226 pKa = 11.84ALPGPNPIRR235 pKa = 11.84ARR237 pKa = 11.84QLARR241 pKa = 11.84SLGQEE246 pKa = 3.46EE247 pKa = 5.4PYY249 pKa = 10.39RR250 pKa = 11.84YY251 pKa = 10.31DD252 pKa = 3.46RR253 pKa = 11.84FEE255 pKa = 4.33TEE257 pKa = 3.45HH258 pKa = 6.84GEE260 pKa = 3.95PSFTPKK266 pKa = 10.06QKK268 pKa = 10.36SADD271 pKa = 3.5YY272 pKa = 10.32VEE274 pKa = 5.85RR275 pKa = 11.84YY276 pKa = 7.48EE277 pKa = 4.4PPGGAYY283 pKa = 9.39QRR285 pKa = 11.84NTPDD289 pKa = 2.53WMLPLILGLYY299 pKa = 10.37GDD301 pKa = 5.24ISPSWGDD308 pKa = 3.14TIEE311 pKa = 4.1RR312 pKa = 11.84VEE314 pKa = 4.13EE315 pKa = 4.13EE316 pKa = 4.32EE317 pKa = 5.02KK318 pKa = 11.11EE319 pKa = 4.24EE320 pKa = 4.15EE321 pKa = 4.85DD322 pKa = 4.14GPKK325 pKa = 9.92KK326 pKa = 10.44KK327 pKa = 10.28KK328 pKa = 10.2LRR330 pKa = 11.84TVRR333 pKa = 11.84GPKK336 pKa = 8.95TNNKK340 pKa = 8.73RR341 pKa = 11.84RR342 pKa = 11.84NRR344 pKa = 11.84STQRR348 pKa = 11.84PNRR351 pKa = 11.84PRR353 pKa = 4.21
Molecular weight: 39.95 kDa
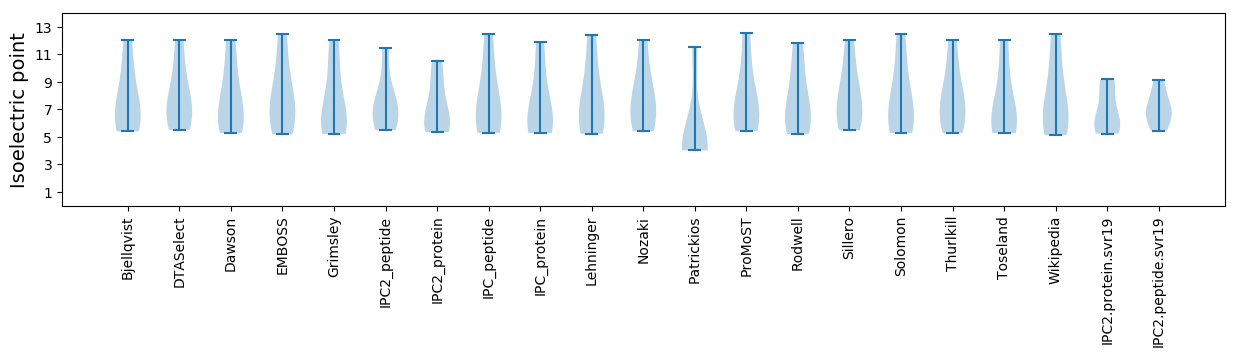
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6DZZ9|B6DZZ9_9POLY Minor capsid protein OS=Myotis polyomavirus VM-2008 OX=563775 PE=3 SV=1
MM1 pKa = 8.07RR2 pKa = 11.84GRR4 pKa = 11.84SSWYY8 pKa = 10.04RR9 pKa = 11.84RR10 pKa = 11.84WKK12 pKa = 10.42DD13 pKa = 3.15LLKK16 pKa = 10.67IFVNLNKK23 pKa = 10.21RR24 pKa = 11.84SGAPPRR30 pKa = 3.92
MM1 pKa = 8.07RR2 pKa = 11.84GRR4 pKa = 11.84SSWYY8 pKa = 10.04RR9 pKa = 11.84RR10 pKa = 11.84WKK12 pKa = 10.42DD13 pKa = 3.15LLKK16 pKa = 10.67IFVNLNKK23 pKa = 10.21RR24 pKa = 11.84SGAPPRR30 pKa = 3.92
Molecular weight: 3.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1809 |
30 |
670 |
301.5 |
34.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.91 ± 1.051 | 1.879 ± 0.742 |
5.694 ± 0.562 | 7.463 ± 0.624 |
4.643 ± 0.555 | 5.804 ± 0.623 |
1.658 ± 0.191 | 5.086 ± 0.108 |
5.915 ± 1.028 | 9.121 ± 0.322 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.432 ± 0.208 | 4.422 ± 0.232 |
6.025 ± 0.73 | 4.367 ± 0.564 |
7.186 ± 1.328 | 5.694 ± 0.33 |
5.196 ± 0.232 | 4.865 ± 0.761 |
1.382 ± 0.253 | 4.256 ± 0.613 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
