
Eubacteriaceae bacterium CHKCI004
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; unclassified Eubacteriaceae
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
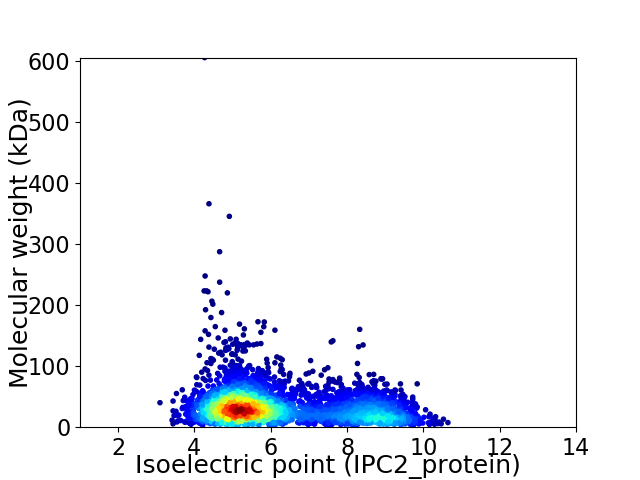
Virtual 2D-PAGE plot for 3682 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A143XJ95|A0A143XJ95_9FIRM Putative acetyltransferase OS=Eubacteriaceae bacterium CHKCI004 OX=1780380 GN=BN3660_00497 PE=4 SV=1
MM1 pKa = 8.15SEE3 pKa = 3.86FDD5 pKa = 3.95YY6 pKa = 11.66NNTNDD11 pKa = 4.65GLNNDD16 pKa = 4.05NTDD19 pKa = 3.31DD20 pKa = 4.57SLNHH24 pKa = 6.94DD25 pKa = 3.78PAGNDD30 pKa = 3.89SLNDD34 pKa = 3.47DD35 pKa = 4.71AGWNKK40 pKa = 10.42PGDD43 pKa = 4.93DD44 pKa = 3.73IDD46 pKa = 4.01TAASEE51 pKa = 4.4EE52 pKa = 4.57TGTSSYY58 pKa = 10.91EE59 pKa = 3.46PAAADD64 pKa = 3.47TAPADD69 pKa = 3.67GTYY72 pKa = 10.11HH73 pKa = 6.17YY74 pKa = 11.16VGDD77 pKa = 4.22PYY79 pKa = 10.97RR80 pKa = 11.84EE81 pKa = 3.98EE82 pKa = 4.53SGTASEE88 pKa = 4.9DD89 pKa = 3.25GRR91 pKa = 11.84NPRR94 pKa = 11.84YY95 pKa = 9.76RR96 pKa = 11.84YY97 pKa = 9.62QGQQNAQYY105 pKa = 10.38NNSFSEE111 pKa = 4.63GSWQGDD117 pKa = 3.54GTSADD122 pKa = 3.8SQQDD126 pKa = 3.16AQYY129 pKa = 11.51NGGAYY134 pKa = 10.08NEE136 pKa = 4.73SSQQYY141 pKa = 8.65YY142 pKa = 10.96SSIPDD147 pKa = 3.46EE148 pKa = 4.39PDD150 pKa = 2.78KK151 pKa = 11.4KK152 pKa = 10.41RR153 pKa = 11.84GGMGMRR159 pKa = 11.84IAKK162 pKa = 9.65IAGAALLFGIVAGVGFAGVSIARR185 pKa = 11.84DD186 pKa = 3.45RR187 pKa = 11.84LIPSATEE194 pKa = 3.72RR195 pKa = 11.84VQTTTVQNNGSDD207 pKa = 3.98DD208 pKa = 4.0SSSSSASAQDD218 pKa = 3.0ISNVVDD224 pKa = 3.51AVMPSVVSITSTTQVSNYY242 pKa = 9.48FFGTQEE248 pKa = 4.05SEE250 pKa = 4.26GAGSGFILAKK260 pKa = 10.07TDD262 pKa = 3.73DD263 pKa = 4.31ALMIATNNHH272 pKa = 4.31VVEE275 pKa = 4.58GANALTVGFSDD286 pKa = 4.06GTTADD291 pKa = 3.39ATIVGTDD298 pKa = 3.12SDD300 pKa = 4.27ADD302 pKa = 3.85LAVISVQASGLSEE315 pKa = 4.22DD316 pKa = 3.78TLSTIKK322 pKa = 10.75VAVLGSSDD330 pKa = 3.64DD331 pKa = 3.93LKK333 pKa = 11.42VGEE336 pKa = 4.26TVIAIGNALGYY347 pKa = 8.84GQSVTTGVVSAKK359 pKa = 10.24DD360 pKa = 3.57RR361 pKa = 11.84EE362 pKa = 4.33VSFTDD367 pKa = 3.38GTMTLLQTDD376 pKa = 3.91AAINPGNSGGVLVNLNGEE394 pKa = 4.33VVGINNAKK402 pKa = 10.68LEE404 pKa = 4.23DD405 pKa = 3.75TSVEE409 pKa = 4.0GMGYY413 pKa = 10.35AIPISTAQGTLEE425 pKa = 3.86TLMNSGSIPEE435 pKa = 4.4GEE437 pKa = 3.8AAYY440 pKa = 10.57LGILGRR446 pKa = 11.84TIDD449 pKa = 3.64STYY452 pKa = 11.08SEE454 pKa = 4.51ALGMPSGVYY463 pKa = 9.41VSQVVEE469 pKa = 4.11GSPAEE474 pKa = 4.13TAGIAAGDD482 pKa = 4.04VITGFEE488 pKa = 4.53GNTVSTMDD496 pKa = 3.67GLKK499 pKa = 10.69DD500 pKa = 4.18RR501 pKa = 11.84ISAQQAGTEE510 pKa = 4.16VQITLQRR517 pKa = 11.84ANQNGTYY524 pKa = 9.95EE525 pKa = 4.26EE526 pKa = 4.16QTVTVTLGKK535 pKa = 10.63EE536 pKa = 3.7SDD538 pKa = 3.91YY539 pKa = 11.54QDD541 pKa = 4.47AEE543 pKa = 4.37TATEE547 pKa = 4.1EE548 pKa = 4.49SADD551 pKa = 3.67TTEE554 pKa = 5.22QDD556 pKa = 3.31SQQYY560 pKa = 7.85GTDD563 pKa = 3.11PYY565 pKa = 11.11EE566 pKa = 4.23YY567 pKa = 10.84YY568 pKa = 10.91NGGNGNDD575 pKa = 4.3YY576 pKa = 10.96YY577 pKa = 11.3SSPYY581 pKa = 10.74DD582 pKa = 3.37YY583 pKa = 11.02FFNN586 pKa = 4.47
MM1 pKa = 8.15SEE3 pKa = 3.86FDD5 pKa = 3.95YY6 pKa = 11.66NNTNDD11 pKa = 4.65GLNNDD16 pKa = 4.05NTDD19 pKa = 3.31DD20 pKa = 4.57SLNHH24 pKa = 6.94DD25 pKa = 3.78PAGNDD30 pKa = 3.89SLNDD34 pKa = 3.47DD35 pKa = 4.71AGWNKK40 pKa = 10.42PGDD43 pKa = 4.93DD44 pKa = 3.73IDD46 pKa = 4.01TAASEE51 pKa = 4.4EE52 pKa = 4.57TGTSSYY58 pKa = 10.91EE59 pKa = 3.46PAAADD64 pKa = 3.47TAPADD69 pKa = 3.67GTYY72 pKa = 10.11HH73 pKa = 6.17YY74 pKa = 11.16VGDD77 pKa = 4.22PYY79 pKa = 10.97RR80 pKa = 11.84EE81 pKa = 3.98EE82 pKa = 4.53SGTASEE88 pKa = 4.9DD89 pKa = 3.25GRR91 pKa = 11.84NPRR94 pKa = 11.84YY95 pKa = 9.76RR96 pKa = 11.84YY97 pKa = 9.62QGQQNAQYY105 pKa = 10.38NNSFSEE111 pKa = 4.63GSWQGDD117 pKa = 3.54GTSADD122 pKa = 3.8SQQDD126 pKa = 3.16AQYY129 pKa = 11.51NGGAYY134 pKa = 10.08NEE136 pKa = 4.73SSQQYY141 pKa = 8.65YY142 pKa = 10.96SSIPDD147 pKa = 3.46EE148 pKa = 4.39PDD150 pKa = 2.78KK151 pKa = 11.4KK152 pKa = 10.41RR153 pKa = 11.84GGMGMRR159 pKa = 11.84IAKK162 pKa = 9.65IAGAALLFGIVAGVGFAGVSIARR185 pKa = 11.84DD186 pKa = 3.45RR187 pKa = 11.84LIPSATEE194 pKa = 3.72RR195 pKa = 11.84VQTTTVQNNGSDD207 pKa = 3.98DD208 pKa = 4.0SSSSSASAQDD218 pKa = 3.0ISNVVDD224 pKa = 3.51AVMPSVVSITSTTQVSNYY242 pKa = 9.48FFGTQEE248 pKa = 4.05SEE250 pKa = 4.26GAGSGFILAKK260 pKa = 10.07TDD262 pKa = 3.73DD263 pKa = 4.31ALMIATNNHH272 pKa = 4.31VVEE275 pKa = 4.58GANALTVGFSDD286 pKa = 4.06GTTADD291 pKa = 3.39ATIVGTDD298 pKa = 3.12SDD300 pKa = 4.27ADD302 pKa = 3.85LAVISVQASGLSEE315 pKa = 4.22DD316 pKa = 3.78TLSTIKK322 pKa = 10.75VAVLGSSDD330 pKa = 3.64DD331 pKa = 3.93LKK333 pKa = 11.42VGEE336 pKa = 4.26TVIAIGNALGYY347 pKa = 8.84GQSVTTGVVSAKK359 pKa = 10.24DD360 pKa = 3.57RR361 pKa = 11.84EE362 pKa = 4.33VSFTDD367 pKa = 3.38GTMTLLQTDD376 pKa = 3.91AAINPGNSGGVLVNLNGEE394 pKa = 4.33VVGINNAKK402 pKa = 10.68LEE404 pKa = 4.23DD405 pKa = 3.75TSVEE409 pKa = 4.0GMGYY413 pKa = 10.35AIPISTAQGTLEE425 pKa = 3.86TLMNSGSIPEE435 pKa = 4.4GEE437 pKa = 3.8AAYY440 pKa = 10.57LGILGRR446 pKa = 11.84TIDD449 pKa = 3.64STYY452 pKa = 11.08SEE454 pKa = 4.51ALGMPSGVYY463 pKa = 9.41VSQVVEE469 pKa = 4.11GSPAEE474 pKa = 4.13TAGIAAGDD482 pKa = 4.04VITGFEE488 pKa = 4.53GNTVSTMDD496 pKa = 3.67GLKK499 pKa = 10.69DD500 pKa = 4.18RR501 pKa = 11.84ISAQQAGTEE510 pKa = 4.16VQITLQRR517 pKa = 11.84ANQNGTYY524 pKa = 9.95EE525 pKa = 4.26EE526 pKa = 4.16QTVTVTLGKK535 pKa = 10.63EE536 pKa = 3.7SDD538 pKa = 3.91YY539 pKa = 11.54QDD541 pKa = 4.47AEE543 pKa = 4.37TATEE547 pKa = 4.1EE548 pKa = 4.49SADD551 pKa = 3.67TTEE554 pKa = 5.22QDD556 pKa = 3.31SQQYY560 pKa = 7.85GTDD563 pKa = 3.11PYY565 pKa = 11.11EE566 pKa = 4.23YY567 pKa = 10.84YY568 pKa = 10.91NGGNGNDD575 pKa = 4.3YY576 pKa = 10.96YY577 pKa = 11.3SSPYY581 pKa = 10.74DD582 pKa = 3.37YY583 pKa = 11.02FFNN586 pKa = 4.47
Molecular weight: 61.14 kDa
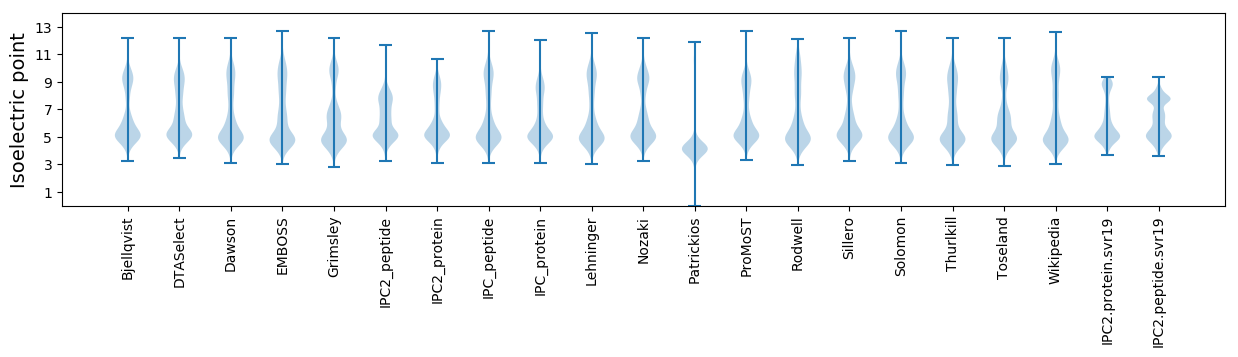
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A143XYV9|A0A143XYV9_9FIRM VWA domain containing CoxE-like protein OS=Eubacteriaceae bacterium CHKCI004 OX=1780380 GN=BN3660_02929 PE=4 SV=1
MM1 pKa = 7.52KK2 pKa = 10.03KK3 pKa = 10.06QRR5 pKa = 11.84VLLGRR10 pKa = 11.84TLAPSAVALATKK22 pKa = 7.91FVCARR27 pKa = 11.84VRR29 pKa = 11.84IPPGEE34 pKa = 3.88LLYY37 pKa = 10.89YY38 pKa = 9.99RR39 pKa = 11.84NSSGISYY46 pKa = 9.88NIKK49 pKa = 10.24KK50 pKa = 10.01LRR52 pKa = 11.84PCKK55 pKa = 9.51RR56 pKa = 11.84DD57 pKa = 3.22EE58 pKa = 4.42VIRR61 pKa = 11.84GSTLIDD67 pKa = 3.57AQTGHH72 pKa = 7.82PLTAVNAWYY81 pKa = 9.66VQRR84 pKa = 11.84SLSMPAGPSHH94 pKa = 6.5CQLPNALSRR103 pKa = 11.84FRR105 pKa = 11.84WNKK108 pKa = 9.55DD109 pKa = 2.73AFSLRR114 pKa = 11.84HH115 pKa = 5.8PLSVSLQNGTLFVLCLFHH133 pKa = 7.84IHH135 pKa = 7.31LSLCNFRR142 pKa = 11.84KK143 pKa = 9.35FVNCFPQKK151 pKa = 9.37MFSVQTISS159 pKa = 2.97
MM1 pKa = 7.52KK2 pKa = 10.03KK3 pKa = 10.06QRR5 pKa = 11.84VLLGRR10 pKa = 11.84TLAPSAVALATKK22 pKa = 7.91FVCARR27 pKa = 11.84VRR29 pKa = 11.84IPPGEE34 pKa = 3.88LLYY37 pKa = 10.89YY38 pKa = 9.99RR39 pKa = 11.84NSSGISYY46 pKa = 9.88NIKK49 pKa = 10.24KK50 pKa = 10.01LRR52 pKa = 11.84PCKK55 pKa = 9.51RR56 pKa = 11.84DD57 pKa = 3.22EE58 pKa = 4.42VIRR61 pKa = 11.84GSTLIDD67 pKa = 3.57AQTGHH72 pKa = 7.82PLTAVNAWYY81 pKa = 9.66VQRR84 pKa = 11.84SLSMPAGPSHH94 pKa = 6.5CQLPNALSRR103 pKa = 11.84FRR105 pKa = 11.84WNKK108 pKa = 9.55DD109 pKa = 2.73AFSLRR114 pKa = 11.84HH115 pKa = 5.8PLSVSLQNGTLFVLCLFHH133 pKa = 7.84IHH135 pKa = 7.31LSLCNFRR142 pKa = 11.84KK143 pKa = 9.35FVNCFPQKK151 pKa = 9.37MFSVQTISS159 pKa = 2.97
Molecular weight: 17.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1165227 |
29 |
5559 |
316.5 |
35.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.492 ± 0.042 | 1.483 ± 0.021 |
5.705 ± 0.038 | 7.912 ± 0.048 |
4.033 ± 0.029 | 7.023 ± 0.043 |
1.786 ± 0.017 | 7.057 ± 0.044 |
6.7 ± 0.042 | 8.835 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.086 ± 0.026 | 4.19 ± 0.03 |
3.333 ± 0.023 | 3.409 ± 0.023 |
4.957 ± 0.043 | 5.871 ± 0.034 |
5.373 ± 0.053 | 6.686 ± 0.036 |
0.909 ± 0.014 | 4.16 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
