
Lake Sarah-associated circular virus-8
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.02
Get precalculated fractions of proteins
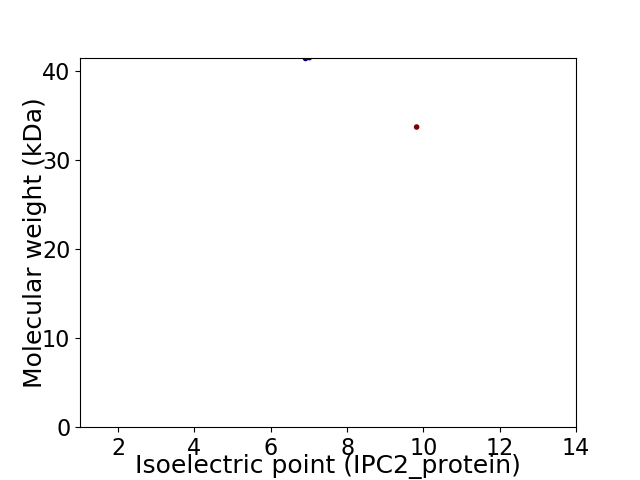
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQK1|A0A140AQK1_9VIRU Replication associated protein (Fragment) OS=Lake Sarah-associated circular virus-8 OX=1685785 PE=3 SV=1
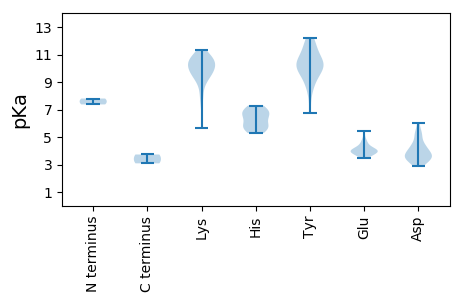
PP1 pKa = 7.8ALRR4 pKa = 11.84ITPTHH9 pKa = 6.09MATNNKK15 pKa = 9.38FSLQAKK21 pKa = 9.6RR22 pKa = 11.84FFLTFPQCPMDD33 pKa = 3.45KK34 pKa = 10.43HH35 pKa = 6.6QVLLWLTGAHH45 pKa = 6.39EE46 pKa = 4.02AHH48 pKa = 5.8QAIVAEE54 pKa = 4.44EE55 pKa = 3.89QHH57 pKa = 6.59EE58 pKa = 4.58DD59 pKa = 3.68GSPHH63 pKa = 5.29LHH65 pKa = 6.94IYY67 pKa = 8.26LTYY70 pKa = 10.16EE71 pKa = 3.49KK72 pKa = 10.46AKK74 pKa = 10.19RR75 pKa = 11.84ISHH78 pKa = 7.04ADD80 pKa = 3.44YY81 pKa = 10.81FDD83 pKa = 4.36LGNPEE88 pKa = 3.96NPEE91 pKa = 4.03QVFHH95 pKa = 7.08PNIQTVKK102 pKa = 10.69NMKK105 pKa = 9.95EE106 pKa = 3.91CVKK109 pKa = 10.92YY110 pKa = 9.24ITKK113 pKa = 9.46TDD115 pKa = 3.63KK116 pKa = 11.03EE117 pKa = 4.27PAQHH121 pKa = 6.55NINYY125 pKa = 9.6RR126 pKa = 11.84DD127 pKa = 3.57IIAGKK132 pKa = 8.59SSKK135 pKa = 10.22FAVIAASVMDD145 pKa = 4.46GKK147 pKa = 11.22SLLEE151 pKa = 4.72INDD154 pKa = 3.72SDD156 pKa = 3.85PGFVLQHH163 pKa = 5.39KK164 pKa = 9.94RR165 pKa = 11.84KK166 pKa = 10.14LEE168 pKa = 4.13EE169 pKa = 3.68YY170 pKa = 9.8QAWIVMKK177 pKa = 10.64KK178 pKa = 9.77PRR180 pKa = 11.84NDD182 pKa = 2.98LAAWYY187 pKa = 6.72PVPVPAFGPEE197 pKa = 3.9YY198 pKa = 10.73QLATWLNANLFAQPRR213 pKa = 11.84RR214 pKa = 11.84IKK216 pKa = 10.66ASQLYY221 pKa = 10.26LFSPPNCGKK230 pKa = 7.34TTLITWLSRR239 pKa = 11.84FCRR242 pKa = 11.84IYY244 pKa = 11.17YY245 pKa = 9.11MPLLEE250 pKa = 5.4DD251 pKa = 5.42FYY253 pKa = 11.64DD254 pKa = 5.11FYY256 pKa = 11.8DD257 pKa = 4.62DD258 pKa = 5.23DD259 pKa = 6.06SYY261 pKa = 12.22DD262 pKa = 3.52LVVIDD267 pKa = 4.36EE268 pKa = 4.9FKK270 pKa = 10.88GQKK273 pKa = 9.49QIQFLNQWLDD283 pKa = 3.6GQPCTVRR290 pKa = 11.84IKK292 pKa = 10.69GGQRR296 pKa = 11.84LKK298 pKa = 10.15TRR300 pKa = 11.84NLPCIILSNWALSTVYY316 pKa = 10.27SAAIEE321 pKa = 4.07KK322 pKa = 10.41SGPDD326 pKa = 3.39VIGPLIARR334 pKa = 11.84LEE336 pKa = 4.06IVNVNKK342 pKa = 10.43FITFYY347 pKa = 11.29DD348 pKa = 4.08PVWVEE353 pKa = 3.96AQNKK357 pKa = 7.97EE358 pKa = 3.96NN359 pKa = 3.73
PP1 pKa = 7.8ALRR4 pKa = 11.84ITPTHH9 pKa = 6.09MATNNKK15 pKa = 9.38FSLQAKK21 pKa = 9.6RR22 pKa = 11.84FFLTFPQCPMDD33 pKa = 3.45KK34 pKa = 10.43HH35 pKa = 6.6QVLLWLTGAHH45 pKa = 6.39EE46 pKa = 4.02AHH48 pKa = 5.8QAIVAEE54 pKa = 4.44EE55 pKa = 3.89QHH57 pKa = 6.59EE58 pKa = 4.58DD59 pKa = 3.68GSPHH63 pKa = 5.29LHH65 pKa = 6.94IYY67 pKa = 8.26LTYY70 pKa = 10.16EE71 pKa = 3.49KK72 pKa = 10.46AKK74 pKa = 10.19RR75 pKa = 11.84ISHH78 pKa = 7.04ADD80 pKa = 3.44YY81 pKa = 10.81FDD83 pKa = 4.36LGNPEE88 pKa = 3.96NPEE91 pKa = 4.03QVFHH95 pKa = 7.08PNIQTVKK102 pKa = 10.69NMKK105 pKa = 9.95EE106 pKa = 3.91CVKK109 pKa = 10.92YY110 pKa = 9.24ITKK113 pKa = 9.46TDD115 pKa = 3.63KK116 pKa = 11.03EE117 pKa = 4.27PAQHH121 pKa = 6.55NINYY125 pKa = 9.6RR126 pKa = 11.84DD127 pKa = 3.57IIAGKK132 pKa = 8.59SSKK135 pKa = 10.22FAVIAASVMDD145 pKa = 4.46GKK147 pKa = 11.22SLLEE151 pKa = 4.72INDD154 pKa = 3.72SDD156 pKa = 3.85PGFVLQHH163 pKa = 5.39KK164 pKa = 9.94RR165 pKa = 11.84KK166 pKa = 10.14LEE168 pKa = 4.13EE169 pKa = 3.68YY170 pKa = 9.8QAWIVMKK177 pKa = 10.64KK178 pKa = 9.77PRR180 pKa = 11.84NDD182 pKa = 2.98LAAWYY187 pKa = 6.72PVPVPAFGPEE197 pKa = 3.9YY198 pKa = 10.73QLATWLNANLFAQPRR213 pKa = 11.84RR214 pKa = 11.84IKK216 pKa = 10.66ASQLYY221 pKa = 10.26LFSPPNCGKK230 pKa = 7.34TTLITWLSRR239 pKa = 11.84FCRR242 pKa = 11.84IYY244 pKa = 11.17YY245 pKa = 9.11MPLLEE250 pKa = 5.4DD251 pKa = 5.42FYY253 pKa = 11.64DD254 pKa = 5.11FYY256 pKa = 11.8DD257 pKa = 4.62DD258 pKa = 5.23DD259 pKa = 6.06SYY261 pKa = 12.22DD262 pKa = 3.52LVVIDD267 pKa = 4.36EE268 pKa = 4.9FKK270 pKa = 10.88GQKK273 pKa = 9.49QIQFLNQWLDD283 pKa = 3.6GQPCTVRR290 pKa = 11.84IKK292 pKa = 10.69GGQRR296 pKa = 11.84LKK298 pKa = 10.15TRR300 pKa = 11.84NLPCIILSNWALSTVYY316 pKa = 10.27SAAIEE321 pKa = 4.07KK322 pKa = 10.41SGPDD326 pKa = 3.39VIGPLIARR334 pKa = 11.84LEE336 pKa = 4.06IVNVNKK342 pKa = 10.43FITFYY347 pKa = 11.29DD348 pKa = 4.08PVWVEE353 pKa = 3.96AQNKK357 pKa = 7.97EE358 pKa = 3.96NN359 pKa = 3.73
Molecular weight: 41.39 kDa
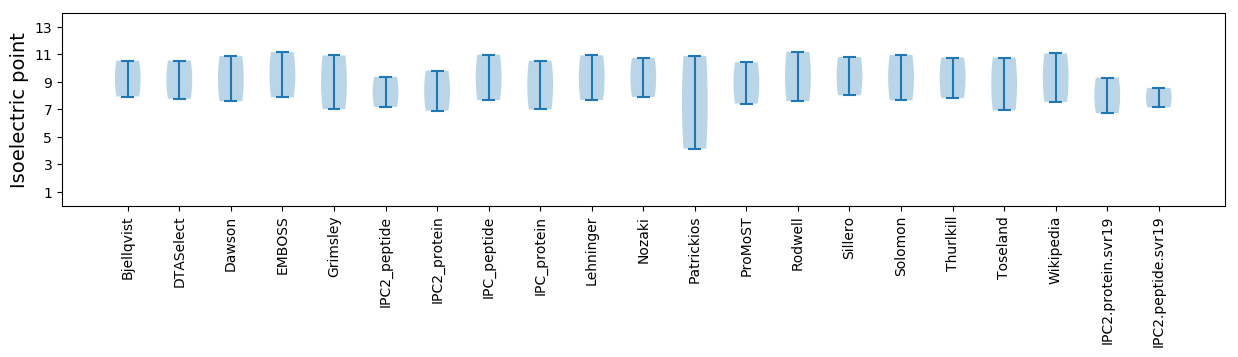
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQK1|A0A140AQK1_9VIRU Replication associated protein (Fragment) OS=Lake Sarah-associated circular virus-8 OX=1685785 PE=3 SV=1
MM1 pKa = 7.37NKK3 pKa = 7.99TWNTGHH9 pKa = 7.29ILPYY13 pKa = 10.43LSAAGTAVQLGWNAWKK29 pKa = 9.67NQGYY33 pKa = 10.0GSYY36 pKa = 9.53EE37 pKa = 3.81LSKK40 pKa = 10.98EE41 pKa = 4.19PEE43 pKa = 3.88KK44 pKa = 11.3KK45 pKa = 9.34MGRR48 pKa = 11.84FRR50 pKa = 11.84QRR52 pKa = 11.84KK53 pKa = 5.63RR54 pKa = 11.84QRR56 pKa = 11.84RR57 pKa = 11.84GRR59 pKa = 11.84FNVRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84KK66 pKa = 10.09SRR68 pKa = 11.84IGGLRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84IRR77 pKa = 11.84KK78 pKa = 9.08INRR81 pKa = 11.84TLFTKK86 pKa = 10.14GVKK89 pKa = 10.09SIEE92 pKa = 3.74VKK94 pKa = 10.82YY95 pKa = 10.62RR96 pKa = 11.84FSTADD101 pKa = 3.23GRR103 pKa = 11.84STSSWSSASTWDD115 pKa = 4.26HH116 pKa = 5.74ITDD119 pKa = 3.47IATGSGPTSRR129 pKa = 11.84VGSKK133 pKa = 10.41VFLRR137 pKa = 11.84HH138 pKa = 5.59LNLRR142 pKa = 11.84MHH144 pKa = 6.01VRR146 pKa = 11.84ASKK149 pKa = 9.04TVPAEE154 pKa = 3.55NEE156 pKa = 3.75QYY158 pKa = 10.34VRR160 pKa = 11.84VMVVRR165 pKa = 11.84HH166 pKa = 5.67SNIQVTPNWSAVSSIPDD183 pKa = 3.57LNILFEE189 pKa = 4.41PALANDD195 pKa = 5.22LDD197 pKa = 4.27YY198 pKa = 11.87QMLPFKK204 pKa = 10.58RR205 pKa = 11.84INNRR209 pKa = 11.84YY210 pKa = 8.66SKK212 pKa = 10.46GFKK215 pKa = 9.81VLYY218 pKa = 10.14SKK220 pKa = 9.71MVKK223 pKa = 9.79VSKK226 pKa = 9.34EE227 pKa = 3.65TGADD231 pKa = 3.31YY232 pKa = 10.34EE233 pKa = 4.11QRR235 pKa = 11.84IWKK238 pKa = 9.7ARR240 pKa = 11.84IPIMKK245 pKa = 8.85PCQWGNTDD253 pKa = 2.9ATGITDD259 pKa = 3.76VGPGQIVIYY268 pKa = 10.05CWDD271 pKa = 3.91NEE273 pKa = 4.13PSALAADD280 pKa = 3.95HH281 pKa = 6.82PLVKK285 pKa = 10.65VGGRR289 pKa = 11.84LSWTDD294 pKa = 3.05VV295 pKa = 3.09
MM1 pKa = 7.37NKK3 pKa = 7.99TWNTGHH9 pKa = 7.29ILPYY13 pKa = 10.43LSAAGTAVQLGWNAWKK29 pKa = 9.67NQGYY33 pKa = 10.0GSYY36 pKa = 9.53EE37 pKa = 3.81LSKK40 pKa = 10.98EE41 pKa = 4.19PEE43 pKa = 3.88KK44 pKa = 11.3KK45 pKa = 9.34MGRR48 pKa = 11.84FRR50 pKa = 11.84QRR52 pKa = 11.84KK53 pKa = 5.63RR54 pKa = 11.84QRR56 pKa = 11.84RR57 pKa = 11.84GRR59 pKa = 11.84FNVRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84KK66 pKa = 10.09SRR68 pKa = 11.84IGGLRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84IRR77 pKa = 11.84KK78 pKa = 9.08INRR81 pKa = 11.84TLFTKK86 pKa = 10.14GVKK89 pKa = 10.09SIEE92 pKa = 3.74VKK94 pKa = 10.82YY95 pKa = 10.62RR96 pKa = 11.84FSTADD101 pKa = 3.23GRR103 pKa = 11.84STSSWSSASTWDD115 pKa = 4.26HH116 pKa = 5.74ITDD119 pKa = 3.47IATGSGPTSRR129 pKa = 11.84VGSKK133 pKa = 10.41VFLRR137 pKa = 11.84HH138 pKa = 5.59LNLRR142 pKa = 11.84MHH144 pKa = 6.01VRR146 pKa = 11.84ASKK149 pKa = 9.04TVPAEE154 pKa = 3.55NEE156 pKa = 3.75QYY158 pKa = 10.34VRR160 pKa = 11.84VMVVRR165 pKa = 11.84HH166 pKa = 5.67SNIQVTPNWSAVSSIPDD183 pKa = 3.57LNILFEE189 pKa = 4.41PALANDD195 pKa = 5.22LDD197 pKa = 4.27YY198 pKa = 11.87QMLPFKK204 pKa = 10.58RR205 pKa = 11.84INNRR209 pKa = 11.84YY210 pKa = 8.66SKK212 pKa = 10.46GFKK215 pKa = 9.81VLYY218 pKa = 10.14SKK220 pKa = 9.71MVKK223 pKa = 9.79VSKK226 pKa = 9.34EE227 pKa = 3.65TGADD231 pKa = 3.31YY232 pKa = 10.34EE233 pKa = 4.11QRR235 pKa = 11.84IWKK238 pKa = 9.7ARR240 pKa = 11.84IPIMKK245 pKa = 8.85PCQWGNTDD253 pKa = 2.9ATGITDD259 pKa = 3.76VGPGQIVIYY268 pKa = 10.05CWDD271 pKa = 3.91NEE273 pKa = 4.13PSALAADD280 pKa = 3.95HH281 pKa = 6.82PLVKK285 pKa = 10.65VGGRR289 pKa = 11.84LSWTDD294 pKa = 3.05VV295 pKa = 3.09
Molecular weight: 33.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
654 |
295 |
359 |
327.0 |
37.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.881 ± 0.453 | 1.223 ± 0.317 |
4.893 ± 0.479 | 4.434 ± 0.607 |
3.976 ± 0.734 | 5.505 ± 1.135 |
2.599 ± 0.329 | 6.422 ± 0.383 |
7.339 ± 0.069 | 7.645 ± 0.897 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.988 ± 0.224 | 5.505 ± 0.047 |
5.657 ± 0.727 | 4.587 ± 0.696 |
6.575 ± 1.891 | 6.269 ± 1.281 |
5.352 ± 0.436 | 6.422 ± 0.602 |
2.752 ± 0.37 | 3.976 ± 0.34 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
