
Pacific flying fox faeces associated circular DNA virus-4
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
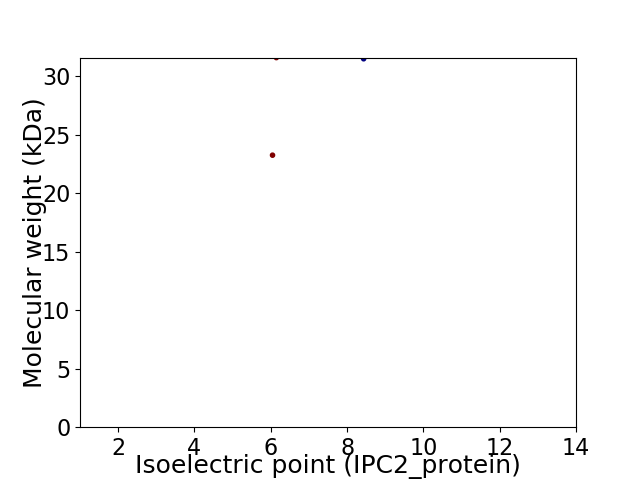
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTT3|A0A140CTT3_9VIRU ATP-dependent helicase Rep OS=Pacific flying fox faeces associated circular DNA virus-4 OX=1796013 PE=3 SV=1
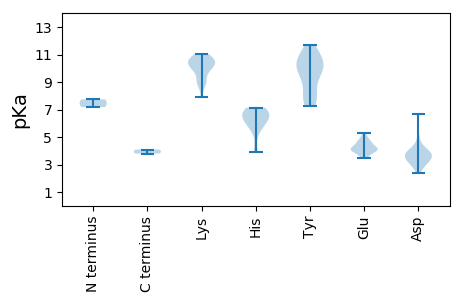
MM1 pKa = 7.18QMTHH5 pKa = 7.13IYY7 pKa = 10.04DD8 pKa = 3.38AEE10 pKa = 3.84IRR12 pKa = 11.84LRR14 pKa = 11.84GDD16 pKa = 3.45GTSDD20 pKa = 3.16AGPTHH25 pKa = 6.79FFFYY29 pKa = 10.52ANSMYY34 pKa = 10.63KK35 pKa = 9.93PWAGVNTNSAEE46 pKa = 4.16GGLSIDD52 pKa = 3.4RR53 pKa = 11.84QPRR56 pKa = 11.84YY57 pKa = 9.29FDD59 pKa = 3.36QMEE62 pKa = 4.75KK63 pKa = 9.96IYY65 pKa = 10.44NHH67 pKa = 6.38HH68 pKa = 5.88VVIGSKK74 pKa = 7.91MTVKK78 pKa = 10.29FSAAGLDD85 pKa = 3.6TAAPTRR91 pKa = 11.84LALRR95 pKa = 11.84QTDD98 pKa = 3.73TNIWDD103 pKa = 3.93SATSNPQMFAEE114 pKa = 4.85FGDD117 pKa = 4.03GKK119 pKa = 10.74DD120 pKa = 3.82RR121 pKa = 11.84ILMATANVPVVMTSKK136 pKa = 10.79YY137 pKa = 8.98SAKK140 pKa = 10.13KK141 pKa = 10.23RR142 pKa = 11.84FGKK145 pKa = 10.75GILANSEE152 pKa = 4.58LKK154 pKa = 10.99CSDD157 pKa = 3.88NGNCSEE163 pKa = 3.63GWYY166 pKa = 10.77YY167 pKa = 11.7NMIVQNMNPNAPNDD181 pKa = 2.79VWAIVRR187 pKa = 11.84IEE189 pKa = 4.33YY190 pKa = 7.9ITVWFEE196 pKa = 4.08IKK198 pKa = 8.83PTNQSTIEE206 pKa = 4.04TT207 pKa = 4.09
MM1 pKa = 7.18QMTHH5 pKa = 7.13IYY7 pKa = 10.04DD8 pKa = 3.38AEE10 pKa = 3.84IRR12 pKa = 11.84LRR14 pKa = 11.84GDD16 pKa = 3.45GTSDD20 pKa = 3.16AGPTHH25 pKa = 6.79FFFYY29 pKa = 10.52ANSMYY34 pKa = 10.63KK35 pKa = 9.93PWAGVNTNSAEE46 pKa = 4.16GGLSIDD52 pKa = 3.4RR53 pKa = 11.84QPRR56 pKa = 11.84YY57 pKa = 9.29FDD59 pKa = 3.36QMEE62 pKa = 4.75KK63 pKa = 9.96IYY65 pKa = 10.44NHH67 pKa = 6.38HH68 pKa = 5.88VVIGSKK74 pKa = 7.91MTVKK78 pKa = 10.29FSAAGLDD85 pKa = 3.6TAAPTRR91 pKa = 11.84LALRR95 pKa = 11.84QTDD98 pKa = 3.73TNIWDD103 pKa = 3.93SATSNPQMFAEE114 pKa = 4.85FGDD117 pKa = 4.03GKK119 pKa = 10.74DD120 pKa = 3.82RR121 pKa = 11.84ILMATANVPVVMTSKK136 pKa = 10.79YY137 pKa = 8.98SAKK140 pKa = 10.13KK141 pKa = 10.23RR142 pKa = 11.84FGKK145 pKa = 10.75GILANSEE152 pKa = 4.58LKK154 pKa = 10.99CSDD157 pKa = 3.88NGNCSEE163 pKa = 3.63GWYY166 pKa = 10.77YY167 pKa = 11.7NMIVQNMNPNAPNDD181 pKa = 2.79VWAIVRR187 pKa = 11.84IEE189 pKa = 4.33YY190 pKa = 7.9ITVWFEE196 pKa = 4.08IKK198 pKa = 8.83PTNQSTIEE206 pKa = 4.04TT207 pKa = 4.09
Molecular weight: 23.26 kDa
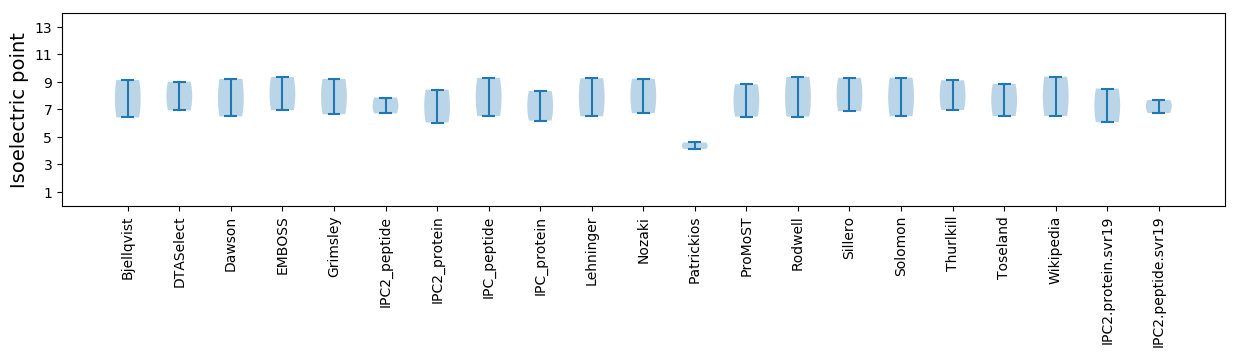
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTT3|A0A140CTT3_9VIRU ATP-dependent helicase Rep OS=Pacific flying fox faeces associated circular DNA virus-4 OX=1796013 PE=3 SV=1
MM1 pKa = 7.74GSQLTQRR8 pKa = 11.84SRR10 pKa = 11.84NFCFTLNNYY19 pKa = 7.83TEE21 pKa = 4.62SDD23 pKa = 3.73TEE25 pKa = 3.95AVKK28 pKa = 10.84AIDD31 pKa = 3.86CKK33 pKa = 10.67YY34 pKa = 8.5MVVGFEE40 pKa = 4.19VGEE43 pKa = 4.34KK44 pKa = 9.04GTPHH48 pKa = 5.6HH49 pKa = 6.08QGYY52 pKa = 9.9IRR54 pKa = 11.84FEE56 pKa = 4.05NARR59 pKa = 11.84SLGATIKK66 pKa = 10.14QLPGKK71 pKa = 8.98PHH73 pKa = 6.8VEE75 pKa = 4.03VAKK78 pKa = 11.04GNASQNIAYY87 pKa = 7.25CTKK90 pKa = 10.37SGVFFEE96 pKa = 4.73KK97 pKa = 9.62GTRR100 pKa = 11.84PIDD103 pKa = 3.67SEE105 pKa = 4.57TCGRR109 pKa = 11.84NEE111 pKa = 3.57KK112 pKa = 10.46RR113 pKa = 11.84RR114 pKa = 11.84WTDD117 pKa = 2.42ARR119 pKa = 11.84AAAKK123 pKa = 9.98EE124 pKa = 4.02GRR126 pKa = 11.84FDD128 pKa = 6.69DD129 pKa = 4.42IPDD132 pKa = 5.01DD133 pKa = 3.2IYY135 pKa = 10.76MRR137 pKa = 11.84HH138 pKa = 3.91VHH140 pKa = 6.52AIKK143 pKa = 10.31RR144 pKa = 11.84IRR146 pKa = 11.84MEE148 pKa = 5.27DD149 pKa = 3.46GPKK152 pKa = 9.09PTDD155 pKa = 3.4LEE157 pKa = 4.2PRR159 pKa = 11.84DD160 pKa = 4.08TYY162 pKa = 11.35GLWIYY167 pKa = 9.66GPPGTGKK174 pKa = 9.65SHH176 pKa = 6.67FVRR179 pKa = 11.84TNFRR183 pKa = 11.84DD184 pKa = 3.74HH185 pKa = 6.8YY186 pKa = 11.0IKK188 pKa = 10.68GANKK192 pKa = 8.73WWTGYY197 pKa = 10.12CGQKK201 pKa = 10.41YY202 pKa = 7.87VVIDD206 pKa = 3.96EE207 pKa = 5.05LSPASGPEE215 pKa = 3.44LSQYY219 pKa = 7.38MKK221 pKa = 10.45KK222 pKa = 9.26WADD225 pKa = 3.0RR226 pKa = 11.84WSFEE230 pKa = 4.43AEE232 pKa = 4.14TKK234 pKa = 10.45GGNSIIRR241 pKa = 11.84PYY243 pKa = 9.94MIIVTSNYY251 pKa = 9.8SIEE254 pKa = 4.27EE255 pKa = 3.93VFSRR259 pKa = 11.84VDD261 pKa = 2.94AQAIKK266 pKa = 10.65RR267 pKa = 11.84RR268 pKa = 11.84FTEE271 pKa = 3.99IKK273 pKa = 10.04FQQ275 pKa = 3.8
MM1 pKa = 7.74GSQLTQRR8 pKa = 11.84SRR10 pKa = 11.84NFCFTLNNYY19 pKa = 7.83TEE21 pKa = 4.62SDD23 pKa = 3.73TEE25 pKa = 3.95AVKK28 pKa = 10.84AIDD31 pKa = 3.86CKK33 pKa = 10.67YY34 pKa = 8.5MVVGFEE40 pKa = 4.19VGEE43 pKa = 4.34KK44 pKa = 9.04GTPHH48 pKa = 5.6HH49 pKa = 6.08QGYY52 pKa = 9.9IRR54 pKa = 11.84FEE56 pKa = 4.05NARR59 pKa = 11.84SLGATIKK66 pKa = 10.14QLPGKK71 pKa = 8.98PHH73 pKa = 6.8VEE75 pKa = 4.03VAKK78 pKa = 11.04GNASQNIAYY87 pKa = 7.25CTKK90 pKa = 10.37SGVFFEE96 pKa = 4.73KK97 pKa = 9.62GTRR100 pKa = 11.84PIDD103 pKa = 3.67SEE105 pKa = 4.57TCGRR109 pKa = 11.84NEE111 pKa = 3.57KK112 pKa = 10.46RR113 pKa = 11.84RR114 pKa = 11.84WTDD117 pKa = 2.42ARR119 pKa = 11.84AAAKK123 pKa = 9.98EE124 pKa = 4.02GRR126 pKa = 11.84FDD128 pKa = 6.69DD129 pKa = 4.42IPDD132 pKa = 5.01DD133 pKa = 3.2IYY135 pKa = 10.76MRR137 pKa = 11.84HH138 pKa = 3.91VHH140 pKa = 6.52AIKK143 pKa = 10.31RR144 pKa = 11.84IRR146 pKa = 11.84MEE148 pKa = 5.27DD149 pKa = 3.46GPKK152 pKa = 9.09PTDD155 pKa = 3.4LEE157 pKa = 4.2PRR159 pKa = 11.84DD160 pKa = 4.08TYY162 pKa = 11.35GLWIYY167 pKa = 9.66GPPGTGKK174 pKa = 9.65SHH176 pKa = 6.67FVRR179 pKa = 11.84TNFRR183 pKa = 11.84DD184 pKa = 3.74HH185 pKa = 6.8YY186 pKa = 11.0IKK188 pKa = 10.68GANKK192 pKa = 8.73WWTGYY197 pKa = 10.12CGQKK201 pKa = 10.41YY202 pKa = 7.87VVIDD206 pKa = 3.96EE207 pKa = 5.05LSPASGPEE215 pKa = 3.44LSQYY219 pKa = 7.38MKK221 pKa = 10.45KK222 pKa = 9.26WADD225 pKa = 3.0RR226 pKa = 11.84WSFEE230 pKa = 4.43AEE232 pKa = 4.14TKK234 pKa = 10.45GGNSIIRR241 pKa = 11.84PYY243 pKa = 9.94MIIVTSNYY251 pKa = 9.8SIEE254 pKa = 4.27EE255 pKa = 3.93VFSRR259 pKa = 11.84VDD261 pKa = 2.94AQAIKK266 pKa = 10.65RR267 pKa = 11.84RR268 pKa = 11.84FTEE271 pKa = 3.99IKK273 pKa = 10.04FQQ275 pKa = 3.8
Molecular weight: 31.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
482 |
207 |
275 |
241.0 |
27.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.469 ± 0.783 | 1.452 ± 0.31 |
5.602 ± 0.125 | 5.809 ± 0.932 |
4.564 ± 0.138 | 7.676 ± 0.583 |
2.282 ± 0.223 | 6.846 ± 0.053 |
6.639 ± 0.845 | 3.32 ± 0.348 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.32 ± 0.964 | 5.602 ± 1.358 |
4.564 ± 0.138 | 3.32 ± 0.04 |
6.224 ± 1.197 | 6.224 ± 0.344 |
7.054 ± 0.431 | 5.187 ± 0.081 |
2.282 ± 0.085 | 4.564 ± 0.138 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
