
Dromedary stool-associated circular ssDNA virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
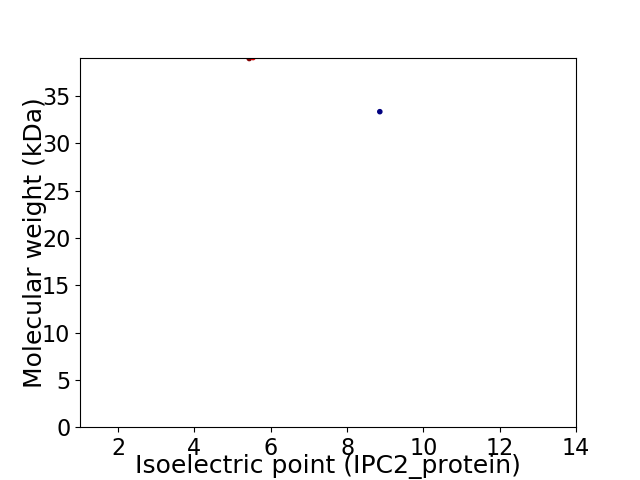
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1ENW9|A0A0A1ENW9_9CIRC ATP-dependent helicase Rep OS=Dromedary stool-associated circular ssDNA virus OX=1574422 GN=rep PE=3 SV=1
MM1 pKa = 7.4SRR3 pKa = 11.84GKK5 pKa = 9.91RR6 pKa = 11.84WCFTINNYY14 pKa = 7.61TEE16 pKa = 4.63EE17 pKa = 4.18DD18 pKa = 3.23TSRR21 pKa = 11.84CEE23 pKa = 5.35RR24 pKa = 11.84IDD26 pKa = 3.6CEE28 pKa = 4.08YY29 pKa = 11.25LVFGKK34 pKa = 10.53EE35 pKa = 3.65IGEE38 pKa = 4.36EE39 pKa = 3.9EE40 pKa = 4.49HH41 pKa = 6.59TPHH44 pKa = 6.23LQGFIVFKK52 pKa = 10.63NRR54 pKa = 11.84KK55 pKa = 5.28TFNVVKK61 pKa = 10.65RR62 pKa = 11.84IIGEE66 pKa = 3.92NAHH69 pKa = 7.15IEE71 pKa = 4.17IARR74 pKa = 11.84GTVKK78 pKa = 10.3EE79 pKa = 3.77ASEE82 pKa = 4.31YY83 pKa = 10.17CKK85 pKa = 10.76KK86 pKa = 10.31EE87 pKa = 3.43GQYY90 pKa = 10.2FEE92 pKa = 5.42KK93 pKa = 11.22GEE95 pKa = 4.12LPPEE99 pKa = 3.92QNEE102 pKa = 4.34RR103 pKa = 11.84GGQATKK109 pKa = 10.6RR110 pKa = 11.84KK111 pKa = 9.33WEE113 pKa = 4.1EE114 pKa = 3.43TLKK117 pKa = 10.72AAKK120 pKa = 9.77EE121 pKa = 4.03GRR123 pKa = 11.84FDD125 pKa = 6.41DD126 pKa = 4.33IAPDD130 pKa = 3.58LYY132 pKa = 10.43IRR134 pKa = 11.84YY135 pKa = 9.24RR136 pKa = 11.84SSLKK140 pKa = 10.43AIYY143 pKa = 9.88QEE145 pKa = 4.13EE146 pKa = 4.55VNKK149 pKa = 8.49NTKK152 pKa = 10.11EE153 pKa = 3.79ITDD156 pKa = 4.15FDD158 pKa = 4.79LKK160 pKa = 11.35GHH162 pKa = 6.57FYY164 pKa = 10.24WIYY167 pKa = 10.65GPTGTGKK174 pKa = 9.65SHH176 pKa = 8.12LARR179 pKa = 11.84TMAASVDD186 pKa = 4.12PDD188 pKa = 3.71TPPYY192 pKa = 10.83LKK194 pKa = 10.66GLNKK198 pKa = 8.92WWSGYY203 pKa = 10.22KK204 pKa = 8.06MQKK207 pKa = 10.36AVIIEE212 pKa = 4.02EE213 pKa = 4.5ANPEE217 pKa = 4.17TCKK220 pKa = 10.94YY221 pKa = 9.79LAPLFKK227 pKa = 10.1QWCDD231 pKa = 2.7KK232 pKa = 10.78WPFTAEE238 pKa = 4.02TKK240 pKa = 10.6GGSFEE245 pKa = 4.5HH246 pKa = 7.43GIRR249 pKa = 11.84PQYY252 pKa = 10.99IFITSNYY259 pKa = 9.41SINEE263 pKa = 4.16CFPDD267 pKa = 4.32PNDD270 pKa = 3.61SEE272 pKa = 4.25PMKK275 pKa = 10.42RR276 pKa = 11.84RR277 pKa = 11.84CHH279 pKa = 6.08EE280 pKa = 3.88FFKK283 pKa = 10.66EE284 pKa = 4.89NKK286 pKa = 9.91DD287 pKa = 3.2SWIPLYY293 pKa = 10.38TDD295 pKa = 3.69NDD297 pKa = 4.04TQTFPIEE304 pKa = 4.08PVHH307 pKa = 6.91LGLNDD312 pKa = 3.43KK313 pKa = 10.51PIDD316 pKa = 3.48IGTQEE321 pKa = 4.52MPLLTPEE328 pKa = 4.94LEE330 pKa = 4.24VLEE333 pKa = 5.22DD334 pKa = 3.71GG335 pKa = 5.14
MM1 pKa = 7.4SRR3 pKa = 11.84GKK5 pKa = 9.91RR6 pKa = 11.84WCFTINNYY14 pKa = 7.61TEE16 pKa = 4.63EE17 pKa = 4.18DD18 pKa = 3.23TSRR21 pKa = 11.84CEE23 pKa = 5.35RR24 pKa = 11.84IDD26 pKa = 3.6CEE28 pKa = 4.08YY29 pKa = 11.25LVFGKK34 pKa = 10.53EE35 pKa = 3.65IGEE38 pKa = 4.36EE39 pKa = 3.9EE40 pKa = 4.49HH41 pKa = 6.59TPHH44 pKa = 6.23LQGFIVFKK52 pKa = 10.63NRR54 pKa = 11.84KK55 pKa = 5.28TFNVVKK61 pKa = 10.65RR62 pKa = 11.84IIGEE66 pKa = 3.92NAHH69 pKa = 7.15IEE71 pKa = 4.17IARR74 pKa = 11.84GTVKK78 pKa = 10.3EE79 pKa = 3.77ASEE82 pKa = 4.31YY83 pKa = 10.17CKK85 pKa = 10.76KK86 pKa = 10.31EE87 pKa = 3.43GQYY90 pKa = 10.2FEE92 pKa = 5.42KK93 pKa = 11.22GEE95 pKa = 4.12LPPEE99 pKa = 3.92QNEE102 pKa = 4.34RR103 pKa = 11.84GGQATKK109 pKa = 10.6RR110 pKa = 11.84KK111 pKa = 9.33WEE113 pKa = 4.1EE114 pKa = 3.43TLKK117 pKa = 10.72AAKK120 pKa = 9.77EE121 pKa = 4.03GRR123 pKa = 11.84FDD125 pKa = 6.41DD126 pKa = 4.33IAPDD130 pKa = 3.58LYY132 pKa = 10.43IRR134 pKa = 11.84YY135 pKa = 9.24RR136 pKa = 11.84SSLKK140 pKa = 10.43AIYY143 pKa = 9.88QEE145 pKa = 4.13EE146 pKa = 4.55VNKK149 pKa = 8.49NTKK152 pKa = 10.11EE153 pKa = 3.79ITDD156 pKa = 4.15FDD158 pKa = 4.79LKK160 pKa = 11.35GHH162 pKa = 6.57FYY164 pKa = 10.24WIYY167 pKa = 10.65GPTGTGKK174 pKa = 9.65SHH176 pKa = 8.12LARR179 pKa = 11.84TMAASVDD186 pKa = 4.12PDD188 pKa = 3.71TPPYY192 pKa = 10.83LKK194 pKa = 10.66GLNKK198 pKa = 8.92WWSGYY203 pKa = 10.22KK204 pKa = 8.06MQKK207 pKa = 10.36AVIIEE212 pKa = 4.02EE213 pKa = 4.5ANPEE217 pKa = 4.17TCKK220 pKa = 10.94YY221 pKa = 9.79LAPLFKK227 pKa = 10.1QWCDD231 pKa = 2.7KK232 pKa = 10.78WPFTAEE238 pKa = 4.02TKK240 pKa = 10.6GGSFEE245 pKa = 4.5HH246 pKa = 7.43GIRR249 pKa = 11.84PQYY252 pKa = 10.99IFITSNYY259 pKa = 9.41SINEE263 pKa = 4.16CFPDD267 pKa = 4.32PNDD270 pKa = 3.61SEE272 pKa = 4.25PMKK275 pKa = 10.42RR276 pKa = 11.84RR277 pKa = 11.84CHH279 pKa = 6.08EE280 pKa = 3.88FFKK283 pKa = 10.66EE284 pKa = 4.89NKK286 pKa = 9.91DD287 pKa = 3.2SWIPLYY293 pKa = 10.38TDD295 pKa = 3.69NDD297 pKa = 4.04TQTFPIEE304 pKa = 4.08PVHH307 pKa = 6.91LGLNDD312 pKa = 3.43KK313 pKa = 10.51PIDD316 pKa = 3.48IGTQEE321 pKa = 4.52MPLLTPEE328 pKa = 4.94LEE330 pKa = 4.24VLEE333 pKa = 5.22DD334 pKa = 3.71GG335 pKa = 5.14
Molecular weight: 38.95 kDa
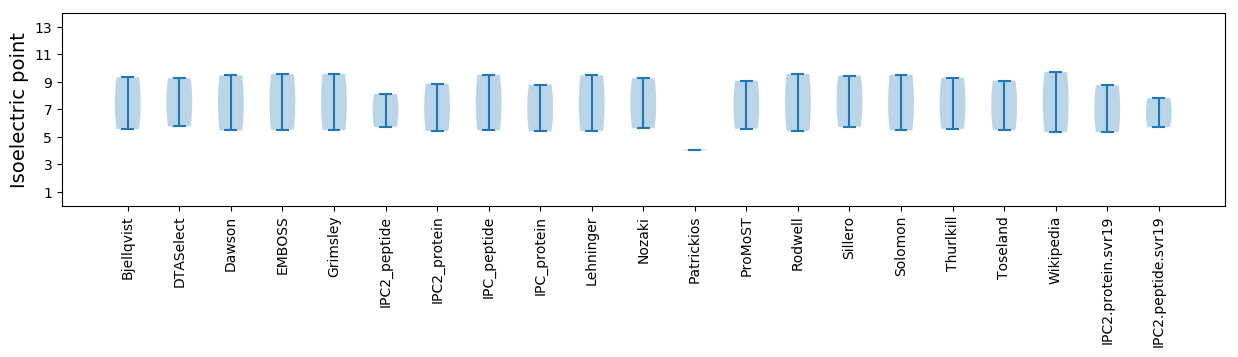
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1ENW9|A0A0A1ENW9_9CIRC ATP-dependent helicase Rep OS=Dromedary stool-associated circular ssDNA virus OX=1574422 GN=rep PE=3 SV=1
MM1 pKa = 7.82PNTVSFQVGTAYY13 pKa = 9.28TKK15 pKa = 10.17KK16 pKa = 9.71RR17 pKa = 11.84KK18 pKa = 9.63RR19 pKa = 11.84MPGTNNVNKK28 pKa = 10.79AEE30 pKa = 4.04IKK32 pKa = 9.35QWNPRR37 pKa = 11.84SFGEE41 pKa = 4.2GNTYY45 pKa = 10.86NGVNIWNVLDD55 pKa = 3.5KK56 pKa = 10.74TLWSVVNSSSWLPRR70 pKa = 11.84PIYY73 pKa = 10.17MPTQGSGSYY82 pKa = 8.66QRR84 pKa = 11.84IGSKK88 pKa = 10.02IFLKK92 pKa = 9.99YY93 pKa = 10.72LRR95 pKa = 11.84FKK97 pKa = 11.09GYY99 pKa = 10.27ISVFWRR105 pKa = 11.84NLIGVRR111 pKa = 11.84WRR113 pKa = 11.84LRR115 pKa = 11.84LLRR118 pKa = 11.84CDD120 pKa = 3.12NYY122 pKa = 11.0LFKK125 pKa = 10.73EE126 pKa = 4.48VTDD129 pKa = 3.86GTTAQAAIIQYY140 pKa = 11.08LDD142 pKa = 3.27LLKK145 pKa = 10.87NHH147 pKa = 6.43EE148 pKa = 4.69HH149 pKa = 6.14PTSGSEE155 pKa = 3.93EE156 pKa = 3.95WSVASTVWDD165 pKa = 3.49NCRR168 pKa = 11.84HH169 pKa = 5.28NFYY172 pKa = 11.05KK173 pKa = 10.46SVKK176 pKa = 9.88DD177 pKa = 3.67VNDD180 pKa = 3.68RR181 pKa = 11.84NVIRR185 pKa = 11.84SKK187 pKa = 11.0VIASGYY193 pKa = 9.77IPVSAPAEE201 pKa = 4.05LVTLGGALTGNAAGAITLSGYY222 pKa = 7.72NTTYY226 pKa = 10.75DD227 pKa = 3.42INNTRR232 pKa = 11.84YY233 pKa = 10.63AMPLDD238 pKa = 3.8VKK240 pKa = 9.63VKK242 pKa = 10.75CNDD245 pKa = 3.43YY246 pKa = 11.25VDD248 pKa = 5.64DD249 pKa = 4.24SVSYY253 pKa = 10.31YY254 pKa = 10.86YY255 pKa = 10.98VLEE258 pKa = 4.24TDD260 pKa = 3.65CGIGISYY267 pKa = 9.47TSANGAYY274 pKa = 10.14ASLATAGALFQLSFFIRR291 pKa = 11.84GYY293 pKa = 9.32FTDD296 pKa = 3.35SS297 pKa = 2.95
MM1 pKa = 7.82PNTVSFQVGTAYY13 pKa = 9.28TKK15 pKa = 10.17KK16 pKa = 9.71RR17 pKa = 11.84KK18 pKa = 9.63RR19 pKa = 11.84MPGTNNVNKK28 pKa = 10.79AEE30 pKa = 4.04IKK32 pKa = 9.35QWNPRR37 pKa = 11.84SFGEE41 pKa = 4.2GNTYY45 pKa = 10.86NGVNIWNVLDD55 pKa = 3.5KK56 pKa = 10.74TLWSVVNSSSWLPRR70 pKa = 11.84PIYY73 pKa = 10.17MPTQGSGSYY82 pKa = 8.66QRR84 pKa = 11.84IGSKK88 pKa = 10.02IFLKK92 pKa = 9.99YY93 pKa = 10.72LRR95 pKa = 11.84FKK97 pKa = 11.09GYY99 pKa = 10.27ISVFWRR105 pKa = 11.84NLIGVRR111 pKa = 11.84WRR113 pKa = 11.84LRR115 pKa = 11.84LLRR118 pKa = 11.84CDD120 pKa = 3.12NYY122 pKa = 11.0LFKK125 pKa = 10.73EE126 pKa = 4.48VTDD129 pKa = 3.86GTTAQAAIIQYY140 pKa = 11.08LDD142 pKa = 3.27LLKK145 pKa = 10.87NHH147 pKa = 6.43EE148 pKa = 4.69HH149 pKa = 6.14PTSGSEE155 pKa = 3.93EE156 pKa = 3.95WSVASTVWDD165 pKa = 3.49NCRR168 pKa = 11.84HH169 pKa = 5.28NFYY172 pKa = 11.05KK173 pKa = 10.46SVKK176 pKa = 9.88DD177 pKa = 3.67VNDD180 pKa = 3.68RR181 pKa = 11.84NVIRR185 pKa = 11.84SKK187 pKa = 11.0VIASGYY193 pKa = 9.77IPVSAPAEE201 pKa = 4.05LVTLGGALTGNAAGAITLSGYY222 pKa = 7.72NTTYY226 pKa = 10.75DD227 pKa = 3.42INNTRR232 pKa = 11.84YY233 pKa = 10.63AMPLDD238 pKa = 3.8VKK240 pKa = 9.63VKK242 pKa = 10.75CNDD245 pKa = 3.43YY246 pKa = 11.25VDD248 pKa = 5.64DD249 pKa = 4.24SVSYY253 pKa = 10.31YY254 pKa = 10.86YY255 pKa = 10.98VLEE258 pKa = 4.24TDD260 pKa = 3.65CGIGISYY267 pKa = 9.47TSANGAYY274 pKa = 10.14ASLATAGALFQLSFFIRR291 pKa = 11.84GYY293 pKa = 9.32FTDD296 pKa = 3.35SS297 pKa = 2.95
Molecular weight: 33.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
632 |
297 |
335 |
316.0 |
36.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.538 ± 0.821 | 1.899 ± 0.379 |
5.063 ± 0.24 | 7.12 ± 3.04 |
4.43 ± 0.499 | 7.278 ± 0.32 |
1.741 ± 0.502 | 6.329 ± 0.416 |
7.278 ± 1.299 | 6.487 ± 0.632 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.424 ± 0.053 | 6.171 ± 1.08 |
4.905 ± 1.056 | 2.69 ± 0.229 |
5.063 ± 0.222 | 6.171 ± 1.774 |
7.12 ± 0.197 | 5.222 ± 1.732 |
2.532 ± 0.111 | 5.538 ± 0.821 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
