
Citrus leaf rugose virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
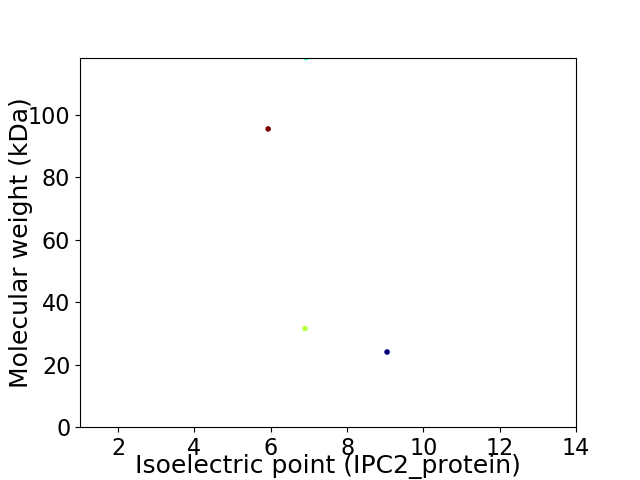
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

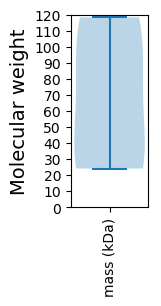
Protein with the lowest isoelectric point:
>tr|Q66108|Q66108_9BROM ATP-dependent helicase OS=Citrus leaf rugose virus OX=37126 PE=3 SV=1
MM1 pKa = 7.75DD2 pKa = 4.28TLLHH6 pKa = 5.93FVKK9 pKa = 10.75FSKK12 pKa = 10.67VLSFLDD18 pKa = 3.62VNATFEE24 pKa = 4.42IPLSDD29 pKa = 3.67EE30 pKa = 3.97QIKK33 pKa = 10.44YY34 pKa = 8.44IQLWMLRR41 pKa = 11.84DD42 pKa = 3.77VVIPAMKK49 pKa = 10.34FSWCGFILLDD59 pKa = 3.72KK60 pKa = 11.44LEE62 pKa = 4.47MEE64 pKa = 5.79LEE66 pKa = 4.38RR67 pKa = 11.84IYY69 pKa = 11.07QNDD72 pKa = 3.83YY73 pKa = 10.7SPPILMHH80 pKa = 6.29VPSQLVTDD88 pKa = 4.58EE89 pKa = 4.5LADD92 pKa = 4.9FIDD95 pKa = 4.14EE96 pKa = 4.32FGEE99 pKa = 4.47VPEE102 pKa = 4.18EE103 pKa = 4.43AYY105 pKa = 10.72LSPEE109 pKa = 3.85EE110 pKa = 4.12RR111 pKa = 11.84EE112 pKa = 4.55KK113 pKa = 10.58IHH115 pKa = 6.98EE116 pKa = 4.1PTFLNHH122 pKa = 5.48EE123 pKa = 4.26HH124 pKa = 6.79WGSEE128 pKa = 3.96SSGSSFLDD136 pKa = 4.35EE137 pKa = 5.26IEE139 pKa = 4.04MLYY142 pKa = 11.05DD143 pKa = 3.39EE144 pKa = 4.66TVKK147 pKa = 11.13DD148 pKa = 4.43DD149 pKa = 3.57VVQDD153 pKa = 3.72VSADD157 pKa = 3.56MPKK160 pKa = 10.39DD161 pKa = 3.75YY162 pKa = 11.15QIMWNDD168 pKa = 3.57GAVDD172 pKa = 3.98AVWSSKK178 pKa = 10.98FEE180 pKa = 4.04VEE182 pKa = 4.53EE183 pKa = 4.07PPKK186 pKa = 10.65PKK188 pKa = 10.38FKK190 pKa = 10.29PQQTDD195 pKa = 4.19RR196 pKa = 11.84ICDD199 pKa = 3.41PVIIQDD205 pKa = 4.7AINDD209 pKa = 3.63IFPFHH214 pKa = 7.83HH215 pKa = 6.62EE216 pKa = 3.71MDD218 pKa = 3.45DD219 pKa = 3.86TYY221 pKa = 11.15FQTWVEE227 pKa = 4.15TQDD230 pKa = 3.07ISLEE234 pKa = 4.18VSKK237 pKa = 10.96CWLDD241 pKa = 3.26ASNFKK246 pKa = 10.96DD247 pKa = 3.78FTKK250 pKa = 10.69GQQTYY255 pKa = 10.28AVPAYY260 pKa = 10.45QSGATSKK267 pKa = 10.53RR268 pKa = 11.84VNTQRR273 pKa = 11.84EE274 pKa = 3.87TLLAVKK280 pKa = 9.35KK281 pKa = 10.43RR282 pKa = 11.84NMNIPEE288 pKa = 4.22LQSTFDD294 pKa = 4.09LDD296 pKa = 4.86AEE298 pKa = 4.38VATCFKK304 pKa = 10.54RR305 pKa = 11.84FCTHH309 pKa = 5.51VVDD312 pKa = 4.41IPRR315 pKa = 11.84SKK317 pKa = 10.69RR318 pKa = 11.84LPQMLGTEE326 pKa = 3.92IQFFSDD332 pKa = 3.37YY333 pKa = 10.92LAGKK337 pKa = 9.45NPPLSEE343 pKa = 4.07YY344 pKa = 10.43QGPLTLQSLDD354 pKa = 3.65KK355 pKa = 10.5YY356 pKa = 8.34MHH358 pKa = 5.5MVKK361 pKa = 9.98TIVKK365 pKa = 8.75PVEE368 pKa = 4.24DD369 pKa = 3.84NSLKK373 pKa = 10.63FEE375 pKa = 4.58RR376 pKa = 11.84PLCATITYY384 pKa = 8.26HH385 pKa = 6.97KK386 pKa = 9.85KK387 pKa = 10.32GIVMQSSPLFLSAMSRR403 pKa = 11.84LFYY406 pKa = 10.61VLKK409 pKa = 10.64SKK411 pKa = 10.19IHH413 pKa = 5.86IPSGKK418 pKa = 7.2WHH420 pKa = 6.3QLFTLDD426 pKa = 4.7AAAFDD431 pKa = 3.55AAKK434 pKa = 9.66WFKK437 pKa = 10.72EE438 pKa = 3.7VDD440 pKa = 3.71FSKK443 pKa = 10.64FDD445 pKa = 3.3KK446 pKa = 11.19SQGEE450 pKa = 4.19LHH452 pKa = 6.89HH453 pKa = 6.79KK454 pKa = 6.68VQKK457 pKa = 10.35MIFDD461 pKa = 4.56LLKK464 pKa = 10.61LPPEE468 pKa = 4.1FVEE471 pKa = 4.27MWFTSHH477 pKa = 5.59EE478 pKa = 3.92RR479 pKa = 11.84SYY481 pKa = 10.12ITDD484 pKa = 3.39RR485 pKa = 11.84DD486 pKa = 3.85TGVGFAVDD494 pKa = 4.05FQRR497 pKa = 11.84RR498 pKa = 11.84TGDD501 pKa = 3.07ANTYY505 pKa = 10.16LGNTLVTLICLARR518 pKa = 11.84VYY520 pKa = 10.32DD521 pKa = 3.84LCDD524 pKa = 3.54PKK526 pKa = 10.22ITFVIASGDD535 pKa = 3.51DD536 pKa = 3.52SLIGSVEE543 pKa = 4.01EE544 pKa = 4.07LPRR547 pKa = 11.84APEE550 pKa = 3.79HH551 pKa = 6.87LFTSLFNFEE560 pKa = 5.0AKK562 pKa = 10.15FPHH565 pKa = 5.93NQPFICSKK573 pKa = 10.38ILVSVDD579 pKa = 3.52LVCGGRR585 pKa = 11.84EE586 pKa = 4.21VIAVPNPAKK595 pKa = 10.65LLIRR599 pKa = 11.84MGRR602 pKa = 11.84RR603 pKa = 11.84DD604 pKa = 3.97CQYY607 pKa = 10.86QALDD611 pKa = 3.78DD612 pKa = 5.92LYY614 pKa = 10.84TSWLDD619 pKa = 3.25VIYY622 pKa = 10.19YY623 pKa = 10.13FRR625 pKa = 11.84DD626 pKa = 3.06SRR628 pKa = 11.84VCEE631 pKa = 4.08KK632 pKa = 10.94VADD635 pKa = 4.0LCAYY639 pKa = 8.84RR640 pKa = 11.84QTRR643 pKa = 11.84RR644 pKa = 11.84SSMYY648 pKa = 10.48LLSALLSLPSCFANKK663 pKa = 10.08KK664 pKa = 9.67KK665 pKa = 10.73FKK667 pKa = 10.18LLCYY671 pKa = 9.95HH672 pKa = 6.46LTEE675 pKa = 4.43EE676 pKa = 4.15EE677 pKa = 4.58CLRR680 pKa = 11.84KK681 pKa = 9.9AKK683 pKa = 10.1PSSITNGHH691 pKa = 5.06VEE693 pKa = 3.77KK694 pKa = 10.15EE695 pKa = 3.9QRR697 pKa = 11.84VNGCRR702 pKa = 11.84DD703 pKa = 2.91SRR705 pKa = 11.84IIQARR710 pKa = 11.84SEE712 pKa = 4.11IPAEE716 pKa = 3.9EE717 pKa = 4.11SRR719 pKa = 11.84RR720 pKa = 11.84IQKK723 pKa = 10.0NSPRR727 pKa = 11.84PDD729 pKa = 3.2KK730 pKa = 11.18GFQAYY735 pKa = 8.14PRR737 pKa = 11.84RR738 pKa = 11.84KK739 pKa = 9.91SRR741 pKa = 11.84GCWMAYY747 pKa = 8.26VPKK750 pKa = 10.18ILQAHH755 pKa = 6.85DD756 pKa = 3.41SSGRR760 pKa = 11.84IPDD763 pKa = 3.29AVTRR767 pKa = 11.84KK768 pKa = 9.53SFSSIRR774 pKa = 11.84GSLGLEE780 pKa = 4.07PDD782 pKa = 4.09PSCGIGVGARR792 pKa = 11.84TTSSFSGKK800 pKa = 6.96NTRR803 pKa = 11.84PRR805 pKa = 11.84HH806 pKa = 5.02RR807 pKa = 11.84FRR809 pKa = 11.84VEE811 pKa = 3.75GVHH814 pKa = 5.51QSSRR818 pKa = 11.84GYY820 pKa = 9.7PADD823 pKa = 3.44LPAYY827 pKa = 9.56NRR829 pKa = 11.84RR830 pKa = 11.84SSS832 pKa = 3.07
MM1 pKa = 7.75DD2 pKa = 4.28TLLHH6 pKa = 5.93FVKK9 pKa = 10.75FSKK12 pKa = 10.67VLSFLDD18 pKa = 3.62VNATFEE24 pKa = 4.42IPLSDD29 pKa = 3.67EE30 pKa = 3.97QIKK33 pKa = 10.44YY34 pKa = 8.44IQLWMLRR41 pKa = 11.84DD42 pKa = 3.77VVIPAMKK49 pKa = 10.34FSWCGFILLDD59 pKa = 3.72KK60 pKa = 11.44LEE62 pKa = 4.47MEE64 pKa = 5.79LEE66 pKa = 4.38RR67 pKa = 11.84IYY69 pKa = 11.07QNDD72 pKa = 3.83YY73 pKa = 10.7SPPILMHH80 pKa = 6.29VPSQLVTDD88 pKa = 4.58EE89 pKa = 4.5LADD92 pKa = 4.9FIDD95 pKa = 4.14EE96 pKa = 4.32FGEE99 pKa = 4.47VPEE102 pKa = 4.18EE103 pKa = 4.43AYY105 pKa = 10.72LSPEE109 pKa = 3.85EE110 pKa = 4.12RR111 pKa = 11.84EE112 pKa = 4.55KK113 pKa = 10.58IHH115 pKa = 6.98EE116 pKa = 4.1PTFLNHH122 pKa = 5.48EE123 pKa = 4.26HH124 pKa = 6.79WGSEE128 pKa = 3.96SSGSSFLDD136 pKa = 4.35EE137 pKa = 5.26IEE139 pKa = 4.04MLYY142 pKa = 11.05DD143 pKa = 3.39EE144 pKa = 4.66TVKK147 pKa = 11.13DD148 pKa = 4.43DD149 pKa = 3.57VVQDD153 pKa = 3.72VSADD157 pKa = 3.56MPKK160 pKa = 10.39DD161 pKa = 3.75YY162 pKa = 11.15QIMWNDD168 pKa = 3.57GAVDD172 pKa = 3.98AVWSSKK178 pKa = 10.98FEE180 pKa = 4.04VEE182 pKa = 4.53EE183 pKa = 4.07PPKK186 pKa = 10.65PKK188 pKa = 10.38FKK190 pKa = 10.29PQQTDD195 pKa = 4.19RR196 pKa = 11.84ICDD199 pKa = 3.41PVIIQDD205 pKa = 4.7AINDD209 pKa = 3.63IFPFHH214 pKa = 7.83HH215 pKa = 6.62EE216 pKa = 3.71MDD218 pKa = 3.45DD219 pKa = 3.86TYY221 pKa = 11.15FQTWVEE227 pKa = 4.15TQDD230 pKa = 3.07ISLEE234 pKa = 4.18VSKK237 pKa = 10.96CWLDD241 pKa = 3.26ASNFKK246 pKa = 10.96DD247 pKa = 3.78FTKK250 pKa = 10.69GQQTYY255 pKa = 10.28AVPAYY260 pKa = 10.45QSGATSKK267 pKa = 10.53RR268 pKa = 11.84VNTQRR273 pKa = 11.84EE274 pKa = 3.87TLLAVKK280 pKa = 9.35KK281 pKa = 10.43RR282 pKa = 11.84NMNIPEE288 pKa = 4.22LQSTFDD294 pKa = 4.09LDD296 pKa = 4.86AEE298 pKa = 4.38VATCFKK304 pKa = 10.54RR305 pKa = 11.84FCTHH309 pKa = 5.51VVDD312 pKa = 4.41IPRR315 pKa = 11.84SKK317 pKa = 10.69RR318 pKa = 11.84LPQMLGTEE326 pKa = 3.92IQFFSDD332 pKa = 3.37YY333 pKa = 10.92LAGKK337 pKa = 9.45NPPLSEE343 pKa = 4.07YY344 pKa = 10.43QGPLTLQSLDD354 pKa = 3.65KK355 pKa = 10.5YY356 pKa = 8.34MHH358 pKa = 5.5MVKK361 pKa = 9.98TIVKK365 pKa = 8.75PVEE368 pKa = 4.24DD369 pKa = 3.84NSLKK373 pKa = 10.63FEE375 pKa = 4.58RR376 pKa = 11.84PLCATITYY384 pKa = 8.26HH385 pKa = 6.97KK386 pKa = 9.85KK387 pKa = 10.32GIVMQSSPLFLSAMSRR403 pKa = 11.84LFYY406 pKa = 10.61VLKK409 pKa = 10.64SKK411 pKa = 10.19IHH413 pKa = 5.86IPSGKK418 pKa = 7.2WHH420 pKa = 6.3QLFTLDD426 pKa = 4.7AAAFDD431 pKa = 3.55AAKK434 pKa = 9.66WFKK437 pKa = 10.72EE438 pKa = 3.7VDD440 pKa = 3.71FSKK443 pKa = 10.64FDD445 pKa = 3.3KK446 pKa = 11.19SQGEE450 pKa = 4.19LHH452 pKa = 6.89HH453 pKa = 6.79KK454 pKa = 6.68VQKK457 pKa = 10.35MIFDD461 pKa = 4.56LLKK464 pKa = 10.61LPPEE468 pKa = 4.1FVEE471 pKa = 4.27MWFTSHH477 pKa = 5.59EE478 pKa = 3.92RR479 pKa = 11.84SYY481 pKa = 10.12ITDD484 pKa = 3.39RR485 pKa = 11.84DD486 pKa = 3.85TGVGFAVDD494 pKa = 4.05FQRR497 pKa = 11.84RR498 pKa = 11.84TGDD501 pKa = 3.07ANTYY505 pKa = 10.16LGNTLVTLICLARR518 pKa = 11.84VYY520 pKa = 10.32DD521 pKa = 3.84LCDD524 pKa = 3.54PKK526 pKa = 10.22ITFVIASGDD535 pKa = 3.51DD536 pKa = 3.52SLIGSVEE543 pKa = 4.01EE544 pKa = 4.07LPRR547 pKa = 11.84APEE550 pKa = 3.79HH551 pKa = 6.87LFTSLFNFEE560 pKa = 5.0AKK562 pKa = 10.15FPHH565 pKa = 5.93NQPFICSKK573 pKa = 10.38ILVSVDD579 pKa = 3.52LVCGGRR585 pKa = 11.84EE586 pKa = 4.21VIAVPNPAKK595 pKa = 10.65LLIRR599 pKa = 11.84MGRR602 pKa = 11.84RR603 pKa = 11.84DD604 pKa = 3.97CQYY607 pKa = 10.86QALDD611 pKa = 3.78DD612 pKa = 5.92LYY614 pKa = 10.84TSWLDD619 pKa = 3.25VIYY622 pKa = 10.19YY623 pKa = 10.13FRR625 pKa = 11.84DD626 pKa = 3.06SRR628 pKa = 11.84VCEE631 pKa = 4.08KK632 pKa = 10.94VADD635 pKa = 4.0LCAYY639 pKa = 8.84RR640 pKa = 11.84QTRR643 pKa = 11.84RR644 pKa = 11.84SSMYY648 pKa = 10.48LLSALLSLPSCFANKK663 pKa = 10.08KK664 pKa = 9.67KK665 pKa = 10.73FKK667 pKa = 10.18LLCYY671 pKa = 9.95HH672 pKa = 6.46LTEE675 pKa = 4.43EE676 pKa = 4.15EE677 pKa = 4.58CLRR680 pKa = 11.84KK681 pKa = 9.9AKK683 pKa = 10.1PSSITNGHH691 pKa = 5.06VEE693 pKa = 3.77KK694 pKa = 10.15EE695 pKa = 3.9QRR697 pKa = 11.84VNGCRR702 pKa = 11.84DD703 pKa = 2.91SRR705 pKa = 11.84IIQARR710 pKa = 11.84SEE712 pKa = 4.11IPAEE716 pKa = 3.9EE717 pKa = 4.11SRR719 pKa = 11.84RR720 pKa = 11.84IQKK723 pKa = 10.0NSPRR727 pKa = 11.84PDD729 pKa = 3.2KK730 pKa = 11.18GFQAYY735 pKa = 8.14PRR737 pKa = 11.84RR738 pKa = 11.84KK739 pKa = 9.91SRR741 pKa = 11.84GCWMAYY747 pKa = 8.26VPKK750 pKa = 10.18ILQAHH755 pKa = 6.85DD756 pKa = 3.41SSGRR760 pKa = 11.84IPDD763 pKa = 3.29AVTRR767 pKa = 11.84KK768 pKa = 9.53SFSSIRR774 pKa = 11.84GSLGLEE780 pKa = 4.07PDD782 pKa = 4.09PSCGIGVGARR792 pKa = 11.84TTSSFSGKK800 pKa = 6.96NTRR803 pKa = 11.84PRR805 pKa = 11.84HH806 pKa = 5.02RR807 pKa = 11.84FRR809 pKa = 11.84VEE811 pKa = 3.75GVHH814 pKa = 5.51QSSRR818 pKa = 11.84GYY820 pKa = 9.7PADD823 pKa = 3.44LPAYY827 pKa = 9.56NRR829 pKa = 11.84RR830 pKa = 11.84SSS832 pKa = 3.07
Molecular weight: 95.49 kDa
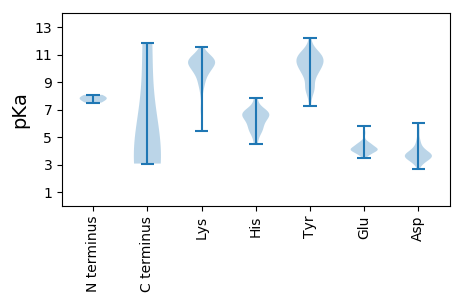
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q66107|Q66107_9BROM RNA-directed RNA polymerase 2a OS=Citrus leaf rugose virus OX=37126 PE=4 SV=1
MM1 pKa = 7.85ANRR4 pKa = 11.84SNAIEE9 pKa = 4.22VNGVWYY15 pKa = 10.55NRR17 pKa = 11.84ADD19 pKa = 3.66NAPVANARR27 pKa = 11.84GRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PTNRR35 pKa = 11.84SRR37 pKa = 11.84NWAQGQRR44 pKa = 11.84SQPQRR49 pKa = 11.84MVVGSMPIDD58 pKa = 4.19TITWKK63 pKa = 10.63SFPGEE68 pKa = 3.8QWHH71 pKa = 6.15EE72 pKa = 3.73FSGFSFPDD80 pKa = 2.71AWGSAKK86 pKa = 9.61IAYY89 pKa = 10.09ASMRR93 pKa = 11.84TEE95 pKa = 3.75LGKK98 pKa = 10.39IKK100 pKa = 10.35SLHH103 pKa = 6.01HH104 pKa = 4.54TTKK107 pKa = 10.27VYY109 pKa = 10.84SVMYY113 pKa = 10.59GFTCKK118 pKa = 10.28ADD120 pKa = 3.74GYY122 pKa = 11.26AGFMDD127 pKa = 4.58NFDD130 pKa = 4.09SANATGPVAPNRR142 pKa = 11.84IRR144 pKa = 11.84VKK146 pKa = 9.99AAKK149 pKa = 9.54YY150 pKa = 9.35CARR153 pKa = 11.84QLVLPPGSTVADD165 pKa = 3.6LRR167 pKa = 11.84DD168 pKa = 3.39NYY170 pKa = 10.82NFVWEE175 pKa = 4.74FDD177 pKa = 3.53AAPAAGTANIISVTKK192 pKa = 9.59FYY194 pKa = 11.26VSTVPLPGIKK204 pKa = 9.85PPPNFLVCEE213 pKa = 4.23MDD215 pKa = 4.76DD216 pKa = 3.58KK217 pKa = 11.87
MM1 pKa = 7.85ANRR4 pKa = 11.84SNAIEE9 pKa = 4.22VNGVWYY15 pKa = 10.55NRR17 pKa = 11.84ADD19 pKa = 3.66NAPVANARR27 pKa = 11.84GRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PTNRR35 pKa = 11.84SRR37 pKa = 11.84NWAQGQRR44 pKa = 11.84SQPQRR49 pKa = 11.84MVVGSMPIDD58 pKa = 4.19TITWKK63 pKa = 10.63SFPGEE68 pKa = 3.8QWHH71 pKa = 6.15EE72 pKa = 3.73FSGFSFPDD80 pKa = 2.71AWGSAKK86 pKa = 9.61IAYY89 pKa = 10.09ASMRR93 pKa = 11.84TEE95 pKa = 3.75LGKK98 pKa = 10.39IKK100 pKa = 10.35SLHH103 pKa = 6.01HH104 pKa = 4.54TTKK107 pKa = 10.27VYY109 pKa = 10.84SVMYY113 pKa = 10.59GFTCKK118 pKa = 10.28ADD120 pKa = 3.74GYY122 pKa = 11.26AGFMDD127 pKa = 4.58NFDD130 pKa = 4.09SANATGPVAPNRR142 pKa = 11.84IRR144 pKa = 11.84VKK146 pKa = 9.99AAKK149 pKa = 9.54YY150 pKa = 9.35CARR153 pKa = 11.84QLVLPPGSTVADD165 pKa = 3.6LRR167 pKa = 11.84DD168 pKa = 3.39NYY170 pKa = 10.82NFVWEE175 pKa = 4.74FDD177 pKa = 3.53AAPAAGTANIISVTKK192 pKa = 9.59FYY194 pKa = 11.26VSTVPLPGIKK204 pKa = 9.85PPPNFLVCEE213 pKa = 4.23MDD215 pKa = 4.76DD216 pKa = 3.58KK217 pKa = 11.87
Molecular weight: 24.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2384 |
217 |
1051 |
596.0 |
67.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.299 ± 0.834 | 1.971 ± 0.179 |
7.005 ± 0.346 | 5.201 ± 0.507 |
4.404 ± 0.623 | 4.488 ± 0.417 |
2.391 ± 0.234 | 5.705 ± 0.296 |
6.04 ± 0.33 | 8.263 ± 0.754 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.894 ± 0.506 | 4.027 ± 0.635 |
5.076 ± 0.527 | 2.894 ± 0.564 |
6.334 ± 0.31 | 8.347 ± 0.203 |
5.663 ± 0.37 | 7.341 ± 0.464 |
1.552 ± 0.21 | 3.104 ± 0.321 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
