
Rhodohalobacter sp. SW132
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; Balneolia; Balneolales; Balneolaceae; Rhodohalobacter; unclassified Rhodohalobacter
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
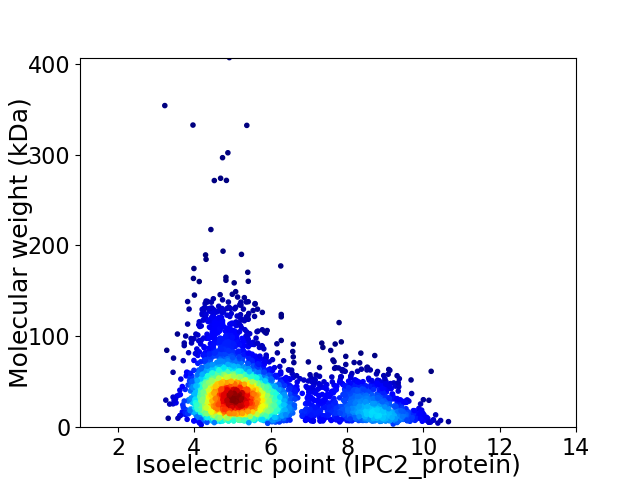
Virtual 2D-PAGE plot for 4092 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A371QIZ6|A0A371QIZ6_9BACT SusC/RagA family TonB-linked outer membrane protein OS=Rhodohalobacter sp. SW132 OX=2293433 GN=DYD21_17495 PE=4 SV=1
MM1 pKa = 7.48RR2 pKa = 11.84QLFTVGIYY10 pKa = 9.92IATLILIASCTKK22 pKa = 10.42EE23 pKa = 3.82NPTNFNMEE31 pKa = 4.3TPQSVAVQDD40 pKa = 4.11SLDD43 pKa = 4.06SNFSPLKK50 pKa = 9.96TYY52 pKa = 10.87INTEE56 pKa = 3.61NSDD59 pKa = 3.96FKK61 pKa = 11.4LGTAVSASAFADD73 pKa = 3.17QGAIFGLVNSNFQEE87 pKa = 4.39VTVHH91 pKa = 6.64DD92 pKa = 4.4MSHH95 pKa = 6.27GAVVQADD102 pKa = 4.1GSHH105 pKa = 7.07DD106 pKa = 4.06LLSISDD112 pKa = 4.75LIGVAEE118 pKa = 3.94EE119 pKa = 4.6AGISVYY125 pKa = 10.97GNALISFEE133 pKa = 4.35NQNEE137 pKa = 4.18AYY139 pKa = 10.48LNDD142 pKa = 5.11LIAPEE147 pKa = 4.39TIEE150 pKa = 3.75VSEE153 pKa = 4.67PSWEE157 pKa = 4.65LISSADD163 pKa = 3.91FEE165 pKa = 4.74TDD167 pKa = 3.22DD168 pKa = 5.14DD169 pKa = 4.96SNYY172 pKa = 9.94EE173 pKa = 4.08ANEE176 pKa = 3.98GADD179 pKa = 3.82LSFTADD185 pKa = 3.18GEE187 pKa = 4.65GANGEE192 pKa = 4.08GRR194 pKa = 11.84ALQIVNAEE202 pKa = 3.92VRR204 pKa = 11.84EE205 pKa = 4.07NDD207 pKa = 2.97WDD209 pKa = 3.84SQFFMTFEE217 pKa = 4.67PNVEE221 pKa = 4.16EE222 pKa = 4.56GDD224 pKa = 3.55QLRR227 pKa = 11.84FVMDD231 pKa = 3.05VRR233 pKa = 11.84AEE235 pKa = 4.04QEE237 pKa = 3.95ASFPTQAHH245 pKa = 6.69DD246 pKa = 4.05APTEE250 pKa = 3.92YY251 pKa = 10.94LHH253 pKa = 7.05WDD255 pKa = 3.88FFGTINATPEE265 pKa = 3.88WSQHH269 pKa = 4.96LMEE272 pKa = 4.56ITVSEE277 pKa = 4.32EE278 pKa = 3.46QASAGTIAFNLGATATTYY296 pKa = 11.08YY297 pKa = 9.66FDD299 pKa = 4.76NMEE302 pKa = 3.45VWYY305 pKa = 10.68YY306 pKa = 8.83NTEE309 pKa = 3.95TGTEE313 pKa = 4.09LVEE316 pKa = 4.01KK317 pKa = 9.91TPEE320 pKa = 3.95EE321 pKa = 4.15KK322 pKa = 10.81EE323 pKa = 4.4EE324 pKa = 3.99ILSTEE329 pKa = 4.03LEE331 pKa = 4.25NWVSTMVSEE340 pKa = 5.23ASYY343 pKa = 10.95VDD345 pKa = 3.0AWDD348 pKa = 3.86VVSGTIEE355 pKa = 4.27GGDD358 pKa = 3.33EE359 pKa = 3.94NSFQLRR365 pKa = 11.84SDD367 pKa = 4.03GGFNWYY373 pKa = 9.67EE374 pKa = 3.71YY375 pKa = 10.44LGEE378 pKa = 4.44DD379 pKa = 4.02FGVQAFQVARR389 pKa = 11.84EE390 pKa = 3.99HH391 pKa = 7.06AGDD394 pKa = 3.83GDD396 pKa = 3.75ILFISDD402 pKa = 3.9YY403 pKa = 11.59GLDD406 pKa = 3.98NLDD409 pKa = 3.88KK410 pKa = 10.25THH412 pKa = 6.62GLINYY417 pKa = 8.95VEE419 pKa = 4.26YY420 pKa = 10.56IEE422 pKa = 5.37NNGAVVDD429 pKa = 5.16GIGTHH434 pKa = 5.69MNININSSRR443 pKa = 11.84QDD445 pKa = 2.87ITEE448 pKa = 3.94MFEE451 pKa = 4.69LLAATDD457 pKa = 4.45KK458 pKa = 10.69IIKK461 pKa = 10.0ISGLNVGLDD470 pKa = 4.2GISAGAASPEE480 pKa = 4.26VYY482 pKa = 9.67EE483 pKa = 4.87AQSEE487 pKa = 4.88MYY489 pKa = 10.46QFVADD494 pKa = 3.74QYY496 pKa = 11.03FSIVPEE502 pKa = 4.0SQRR505 pKa = 11.84YY506 pKa = 8.94GITIWNPLDD515 pKa = 4.46SSDD518 pKa = 4.19NPSGLWTSDD527 pKa = 3.36YY528 pKa = 10.61EE529 pKa = 4.47RR530 pKa = 11.84KK531 pKa = 9.57RR532 pKa = 11.84AYY534 pKa = 10.41AGFAVGLMNGFNSGNN549 pKa = 3.38
MM1 pKa = 7.48RR2 pKa = 11.84QLFTVGIYY10 pKa = 9.92IATLILIASCTKK22 pKa = 10.42EE23 pKa = 3.82NPTNFNMEE31 pKa = 4.3TPQSVAVQDD40 pKa = 4.11SLDD43 pKa = 4.06SNFSPLKK50 pKa = 9.96TYY52 pKa = 10.87INTEE56 pKa = 3.61NSDD59 pKa = 3.96FKK61 pKa = 11.4LGTAVSASAFADD73 pKa = 3.17QGAIFGLVNSNFQEE87 pKa = 4.39VTVHH91 pKa = 6.64DD92 pKa = 4.4MSHH95 pKa = 6.27GAVVQADD102 pKa = 4.1GSHH105 pKa = 7.07DD106 pKa = 4.06LLSISDD112 pKa = 4.75LIGVAEE118 pKa = 3.94EE119 pKa = 4.6AGISVYY125 pKa = 10.97GNALISFEE133 pKa = 4.35NQNEE137 pKa = 4.18AYY139 pKa = 10.48LNDD142 pKa = 5.11LIAPEE147 pKa = 4.39TIEE150 pKa = 3.75VSEE153 pKa = 4.67PSWEE157 pKa = 4.65LISSADD163 pKa = 3.91FEE165 pKa = 4.74TDD167 pKa = 3.22DD168 pKa = 5.14DD169 pKa = 4.96SNYY172 pKa = 9.94EE173 pKa = 4.08ANEE176 pKa = 3.98GADD179 pKa = 3.82LSFTADD185 pKa = 3.18GEE187 pKa = 4.65GANGEE192 pKa = 4.08GRR194 pKa = 11.84ALQIVNAEE202 pKa = 3.92VRR204 pKa = 11.84EE205 pKa = 4.07NDD207 pKa = 2.97WDD209 pKa = 3.84SQFFMTFEE217 pKa = 4.67PNVEE221 pKa = 4.16EE222 pKa = 4.56GDD224 pKa = 3.55QLRR227 pKa = 11.84FVMDD231 pKa = 3.05VRR233 pKa = 11.84AEE235 pKa = 4.04QEE237 pKa = 3.95ASFPTQAHH245 pKa = 6.69DD246 pKa = 4.05APTEE250 pKa = 3.92YY251 pKa = 10.94LHH253 pKa = 7.05WDD255 pKa = 3.88FFGTINATPEE265 pKa = 3.88WSQHH269 pKa = 4.96LMEE272 pKa = 4.56ITVSEE277 pKa = 4.32EE278 pKa = 3.46QASAGTIAFNLGATATTYY296 pKa = 11.08YY297 pKa = 9.66FDD299 pKa = 4.76NMEE302 pKa = 3.45VWYY305 pKa = 10.68YY306 pKa = 8.83NTEE309 pKa = 3.95TGTEE313 pKa = 4.09LVEE316 pKa = 4.01KK317 pKa = 9.91TPEE320 pKa = 3.95EE321 pKa = 4.15KK322 pKa = 10.81EE323 pKa = 4.4EE324 pKa = 3.99ILSTEE329 pKa = 4.03LEE331 pKa = 4.25NWVSTMVSEE340 pKa = 5.23ASYY343 pKa = 10.95VDD345 pKa = 3.0AWDD348 pKa = 3.86VVSGTIEE355 pKa = 4.27GGDD358 pKa = 3.33EE359 pKa = 3.94NSFQLRR365 pKa = 11.84SDD367 pKa = 4.03GGFNWYY373 pKa = 9.67EE374 pKa = 3.71YY375 pKa = 10.44LGEE378 pKa = 4.44DD379 pKa = 4.02FGVQAFQVARR389 pKa = 11.84EE390 pKa = 3.99HH391 pKa = 7.06AGDD394 pKa = 3.83GDD396 pKa = 3.75ILFISDD402 pKa = 3.9YY403 pKa = 11.59GLDD406 pKa = 3.98NLDD409 pKa = 3.88KK410 pKa = 10.25THH412 pKa = 6.62GLINYY417 pKa = 8.95VEE419 pKa = 4.26YY420 pKa = 10.56IEE422 pKa = 5.37NNGAVVDD429 pKa = 5.16GIGTHH434 pKa = 5.69MNININSSRR443 pKa = 11.84QDD445 pKa = 2.87ITEE448 pKa = 3.94MFEE451 pKa = 4.69LLAATDD457 pKa = 4.45KK458 pKa = 10.69IIKK461 pKa = 10.0ISGLNVGLDD470 pKa = 4.2GISAGAASPEE480 pKa = 4.26VYY482 pKa = 9.67EE483 pKa = 4.87AQSEE487 pKa = 4.88MYY489 pKa = 10.46QFVADD494 pKa = 3.74QYY496 pKa = 11.03FSIVPEE502 pKa = 4.0SQRR505 pKa = 11.84YY506 pKa = 8.94GITIWNPLDD515 pKa = 4.46SSDD518 pKa = 4.19NPSGLWTSDD527 pKa = 3.36YY528 pKa = 10.61EE529 pKa = 4.47RR530 pKa = 11.84KK531 pKa = 9.57RR532 pKa = 11.84AYY534 pKa = 10.41AGFAVGLMNGFNSGNN549 pKa = 3.38
Molecular weight: 60.5 kDa
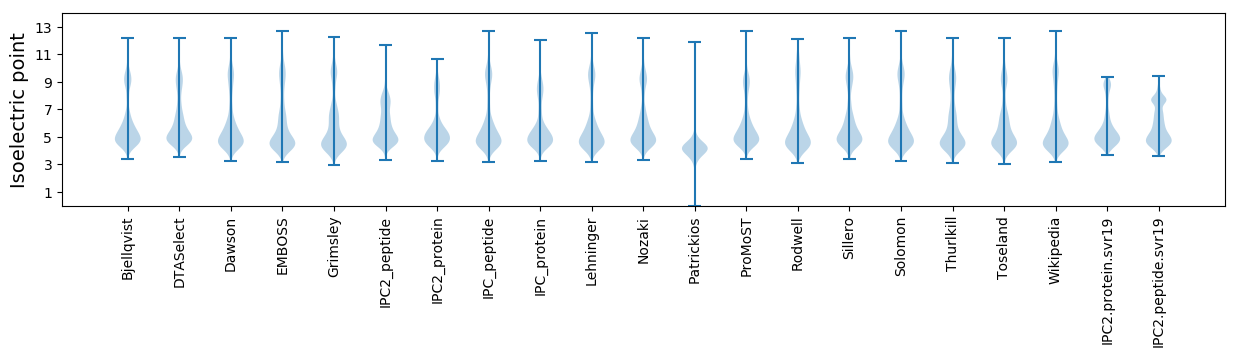
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A371QHH9|A0A371QHH9_9BACT ATP-binding protein OS=Rhodohalobacter sp. SW132 OX=2293433 GN=DYD21_19700 PE=4 SV=1
AA1 pKa = 7.67AWQEE5 pKa = 3.77SGRR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 10.04GSPRR14 pKa = 11.84IYY16 pKa = 10.74NQLRR20 pKa = 11.84KK21 pKa = 10.17DD22 pKa = 3.14GCTASRR28 pKa = 11.84PRR30 pKa = 11.84IARR33 pKa = 11.84LMVKK37 pKa = 9.75MGIASRR43 pKa = 11.84IRR45 pKa = 11.84KK46 pKa = 8.73KK47 pKa = 9.61WIKK50 pKa = 7.15TTHH53 pKa = 5.61STHH56 pKa = 5.79GWPVAANVLDD66 pKa = 4.61RR67 pKa = 11.84NFSPEE72 pKa = 3.48GLGQVWVSDD81 pKa = 3.01ITYY84 pKa = 10.15IRR86 pKa = 11.84GEE88 pKa = 4.19QGWMYY93 pKa = 11.15LL94 pKa = 3.63
AA1 pKa = 7.67AWQEE5 pKa = 3.77SGRR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 10.04GSPRR14 pKa = 11.84IYY16 pKa = 10.74NQLRR20 pKa = 11.84KK21 pKa = 10.17DD22 pKa = 3.14GCTASRR28 pKa = 11.84PRR30 pKa = 11.84IARR33 pKa = 11.84LMVKK37 pKa = 9.75MGIASRR43 pKa = 11.84IRR45 pKa = 11.84KK46 pKa = 8.73KK47 pKa = 9.61WIKK50 pKa = 7.15TTHH53 pKa = 5.61STHH56 pKa = 5.79GWPVAANVLDD66 pKa = 4.61RR67 pKa = 11.84NFSPEE72 pKa = 3.48GLGQVWVSDD81 pKa = 3.01ITYY84 pKa = 10.15IRR86 pKa = 11.84GEE88 pKa = 4.19QGWMYY93 pKa = 11.15LL94 pKa = 3.63
Molecular weight: 10.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1445183 |
22 |
3562 |
353.2 |
39.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.69 ± 0.031 | 0.602 ± 0.011 |
6.312 ± 0.029 | 7.546 ± 0.038 |
4.759 ± 0.029 | 7.194 ± 0.043 |
2.133 ± 0.02 | 7.029 ± 0.032 |
4.928 ± 0.048 | 9.203 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.018 | 4.868 ± 0.028 |
4.06 ± 0.023 | 3.629 ± 0.025 |
5.139 ± 0.034 | 6.903 ± 0.038 |
5.537 ± 0.031 | 6.242 ± 0.027 |
1.288 ± 0.017 | 3.517 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
