
Alistipes sp. CAG:268
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; environmental samples
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
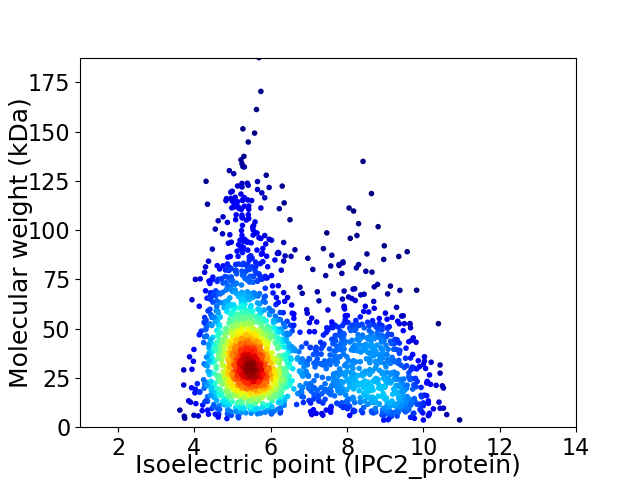
Virtual 2D-PAGE plot for 2200 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
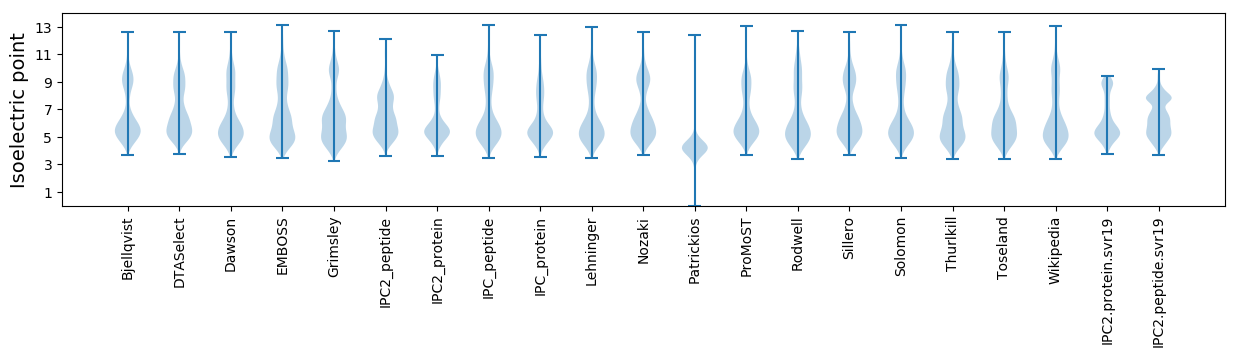
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6VQN4|R6VQN4_9BACT Uncharacterized protein OS=Alistipes sp. CAG:268 OX=1262693 GN=BN576_01836 PE=4 SV=1
MM1 pKa = 7.52KK2 pKa = 9.92IKK4 pKa = 10.27RR5 pKa = 11.84YY6 pKa = 10.79AMMLAATTGLFAACQDD22 pKa = 3.79LDD24 pKa = 3.82EE25 pKa = 4.43MHH27 pKa = 7.04TYY29 pKa = 10.72APEE32 pKa = 4.2DD33 pKa = 3.4VVAPVLHH40 pKa = 6.44NLPEE44 pKa = 4.63EE45 pKa = 4.0IVITSEE51 pKa = 4.4NMGEE55 pKa = 4.3TQVFTWDD62 pKa = 3.24AADD65 pKa = 4.49FGVSTQINYY74 pKa = 10.32SLEE77 pKa = 4.22ASCGEE82 pKa = 4.03GDD84 pKa = 4.08PVVLFADD91 pKa = 3.99LTDD94 pKa = 3.74TSTEE98 pKa = 3.82QPYY101 pKa = 11.0EE102 pKa = 4.27SINTKK107 pKa = 10.08LVYY110 pKa = 10.2DD111 pKa = 4.57AGVEE115 pKa = 4.16PDD117 pKa = 3.93TPTQVNFYY125 pKa = 9.89ISATIGSDD133 pKa = 3.45FEE135 pKa = 4.5KK136 pKa = 10.55FYY138 pKa = 10.7SAPVAVTMTVTKK150 pKa = 10.59AEE152 pKa = 3.92RR153 pKa = 11.84VYY155 pKa = 9.5PTVWVIGDD163 pKa = 3.5YY164 pKa = 11.13CGWNHH169 pKa = 6.47SNSQFLFSFSDD180 pKa = 3.72DD181 pKa = 3.97EE182 pKa = 4.44INYY185 pKa = 10.45EE186 pKa = 3.93GMIDD190 pKa = 4.0FGEE193 pKa = 4.41KK194 pKa = 9.73AANGFKK200 pKa = 10.14LTGIGGWDD208 pKa = 4.05DD209 pKa = 3.54SCNWGTDD216 pKa = 3.49GEE218 pKa = 4.55AEE220 pKa = 4.07APEE223 pKa = 5.08AEE225 pKa = 4.18AASIKK230 pKa = 10.49LISAGNSGNITAYY243 pKa = 9.92SKK245 pKa = 10.6RR246 pKa = 11.84FYY248 pKa = 10.61RR249 pKa = 11.84FAFNRR254 pKa = 11.84EE255 pKa = 3.96SLTLTKK261 pKa = 10.26EE262 pKa = 3.86LSFDD266 pKa = 3.27QLGIVGDD273 pKa = 4.82GVGSWDD279 pKa = 3.48NDD281 pKa = 3.06VVMEE285 pKa = 4.95FDD287 pKa = 3.45TKK289 pKa = 7.84TQRR292 pKa = 11.84FWADD296 pKa = 3.07VEE298 pKa = 4.47LVSGAIKK305 pKa = 10.26FRR307 pKa = 11.84ADD309 pKa = 3.68GAWDD313 pKa = 3.56TSFGSSTEE321 pKa = 3.87GMLDD325 pKa = 3.24GSDD328 pKa = 4.2NIQVPAGNYY337 pKa = 9.48RR338 pKa = 11.84IYY340 pKa = 11.65VNLNNSAAMTYY351 pKa = 9.98EE352 pKa = 4.35LNADD356 pKa = 4.37DD357 pKa = 4.73YY358 pKa = 9.77GQSGGEE364 pKa = 4.03EE365 pKa = 4.13EE366 pKa = 5.17PDD368 pKa = 3.56PEE370 pKa = 4.07TADD373 pKa = 3.47WYY375 pKa = 10.4IHH377 pKa = 5.54GQTVATPDD385 pKa = 3.14WGPTPMEE392 pKa = 4.35SASSNIVAYY401 pKa = 9.46KK402 pKa = 9.92AAGVEE407 pKa = 4.42VAANSQFLFKK417 pKa = 10.94SGDD420 pKa = 3.24EE421 pKa = 4.4SQWLGADD428 pKa = 3.33ASLAGGDD435 pKa = 3.72EE436 pKa = 4.75MYY438 pKa = 10.0TCTVGSAFAVSANKK452 pKa = 9.81VNAVIAEE459 pKa = 4.24AGTYY463 pKa = 9.84DD464 pKa = 3.71YY465 pKa = 11.01WLLPEE470 pKa = 4.81AGRR473 pKa = 11.84AYY475 pKa = 11.1VMAAGVKK482 pKa = 8.97PEE484 pKa = 4.12YY485 pKa = 10.38VAEE488 pKa = 4.08TWGLIGTISGWGDD501 pKa = 3.32LGDD504 pKa = 4.13LSMSEE509 pKa = 3.84QEE511 pKa = 4.51GYY513 pKa = 10.45LVRR516 pKa = 11.84KK517 pKa = 9.63GVALTTADD525 pKa = 3.29EE526 pKa = 4.45FKK528 pKa = 10.8IRR530 pKa = 11.84FNNEE534 pKa = 2.8WDD536 pKa = 3.44DD537 pKa = 3.79SKK539 pKa = 11.94NYY541 pKa = 8.15GTASGGAIDD550 pKa = 4.8INTAVAVVTSGGSQNMKK567 pKa = 9.62VQLDD571 pKa = 3.92GTYY574 pKa = 10.42DD575 pKa = 3.12IYY577 pKa = 11.43FDD579 pKa = 4.38LANSTIYY586 pKa = 11.11VMSPGKK592 pKa = 8.89TPADD596 pKa = 3.63VQQ598 pKa = 3.61
MM1 pKa = 7.52KK2 pKa = 9.92IKK4 pKa = 10.27RR5 pKa = 11.84YY6 pKa = 10.79AMMLAATTGLFAACQDD22 pKa = 3.79LDD24 pKa = 3.82EE25 pKa = 4.43MHH27 pKa = 7.04TYY29 pKa = 10.72APEE32 pKa = 4.2DD33 pKa = 3.4VVAPVLHH40 pKa = 6.44NLPEE44 pKa = 4.63EE45 pKa = 4.0IVITSEE51 pKa = 4.4NMGEE55 pKa = 4.3TQVFTWDD62 pKa = 3.24AADD65 pKa = 4.49FGVSTQINYY74 pKa = 10.32SLEE77 pKa = 4.22ASCGEE82 pKa = 4.03GDD84 pKa = 4.08PVVLFADD91 pKa = 3.99LTDD94 pKa = 3.74TSTEE98 pKa = 3.82QPYY101 pKa = 11.0EE102 pKa = 4.27SINTKK107 pKa = 10.08LVYY110 pKa = 10.2DD111 pKa = 4.57AGVEE115 pKa = 4.16PDD117 pKa = 3.93TPTQVNFYY125 pKa = 9.89ISATIGSDD133 pKa = 3.45FEE135 pKa = 4.5KK136 pKa = 10.55FYY138 pKa = 10.7SAPVAVTMTVTKK150 pKa = 10.59AEE152 pKa = 3.92RR153 pKa = 11.84VYY155 pKa = 9.5PTVWVIGDD163 pKa = 3.5YY164 pKa = 11.13CGWNHH169 pKa = 6.47SNSQFLFSFSDD180 pKa = 3.72DD181 pKa = 3.97EE182 pKa = 4.44INYY185 pKa = 10.45EE186 pKa = 3.93GMIDD190 pKa = 4.0FGEE193 pKa = 4.41KK194 pKa = 9.73AANGFKK200 pKa = 10.14LTGIGGWDD208 pKa = 4.05DD209 pKa = 3.54SCNWGTDD216 pKa = 3.49GEE218 pKa = 4.55AEE220 pKa = 4.07APEE223 pKa = 5.08AEE225 pKa = 4.18AASIKK230 pKa = 10.49LISAGNSGNITAYY243 pKa = 9.92SKK245 pKa = 10.6RR246 pKa = 11.84FYY248 pKa = 10.61RR249 pKa = 11.84FAFNRR254 pKa = 11.84EE255 pKa = 3.96SLTLTKK261 pKa = 10.26EE262 pKa = 3.86LSFDD266 pKa = 3.27QLGIVGDD273 pKa = 4.82GVGSWDD279 pKa = 3.48NDD281 pKa = 3.06VVMEE285 pKa = 4.95FDD287 pKa = 3.45TKK289 pKa = 7.84TQRR292 pKa = 11.84FWADD296 pKa = 3.07VEE298 pKa = 4.47LVSGAIKK305 pKa = 10.26FRR307 pKa = 11.84ADD309 pKa = 3.68GAWDD313 pKa = 3.56TSFGSSTEE321 pKa = 3.87GMLDD325 pKa = 3.24GSDD328 pKa = 4.2NIQVPAGNYY337 pKa = 9.48RR338 pKa = 11.84IYY340 pKa = 11.65VNLNNSAAMTYY351 pKa = 9.98EE352 pKa = 4.35LNADD356 pKa = 4.37DD357 pKa = 4.73YY358 pKa = 9.77GQSGGEE364 pKa = 4.03EE365 pKa = 4.13EE366 pKa = 5.17PDD368 pKa = 3.56PEE370 pKa = 4.07TADD373 pKa = 3.47WYY375 pKa = 10.4IHH377 pKa = 5.54GQTVATPDD385 pKa = 3.14WGPTPMEE392 pKa = 4.35SASSNIVAYY401 pKa = 9.46KK402 pKa = 9.92AAGVEE407 pKa = 4.42VAANSQFLFKK417 pKa = 10.94SGDD420 pKa = 3.24EE421 pKa = 4.4SQWLGADD428 pKa = 3.33ASLAGGDD435 pKa = 3.72EE436 pKa = 4.75MYY438 pKa = 10.0TCTVGSAFAVSANKK452 pKa = 9.81VNAVIAEE459 pKa = 4.24AGTYY463 pKa = 9.84DD464 pKa = 3.71YY465 pKa = 11.01WLLPEE470 pKa = 4.81AGRR473 pKa = 11.84AYY475 pKa = 11.1VMAAGVKK482 pKa = 8.97PEE484 pKa = 4.12YY485 pKa = 10.38VAEE488 pKa = 4.08TWGLIGTISGWGDD501 pKa = 3.32LGDD504 pKa = 4.13LSMSEE509 pKa = 3.84QEE511 pKa = 4.51GYY513 pKa = 10.45LVRR516 pKa = 11.84KK517 pKa = 9.63GVALTTADD525 pKa = 3.29EE526 pKa = 4.45FKK528 pKa = 10.8IRR530 pKa = 11.84FNNEE534 pKa = 2.8WDD536 pKa = 3.44DD537 pKa = 3.79SKK539 pKa = 11.94NYY541 pKa = 8.15GTASGGAIDD550 pKa = 4.8INTAVAVVTSGGSQNMKK567 pKa = 9.62VQLDD571 pKa = 3.92GTYY574 pKa = 10.42DD575 pKa = 3.12IYY577 pKa = 11.43FDD579 pKa = 4.38LANSTIYY586 pKa = 11.11VMSPGKK592 pKa = 8.89TPADD596 pKa = 3.63VQQ598 pKa = 3.61
Molecular weight: 64.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6VCK5|R6VCK5_9BACT Uncharacterized protein OS=Alistipes sp. CAG:268 OX=1262693 GN=BN576_01531 PE=4 SV=1
MM1 pKa = 7.84PNGKK5 pKa = 8.55KK6 pKa = 9.72HH7 pKa = 6.1KK8 pKa = 7.52RR9 pKa = 11.84HH10 pKa = 5.93KK11 pKa = 9.95MATHH15 pKa = 6.02KK16 pKa = 10.34RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.26NRR25 pKa = 11.84HH26 pKa = 4.71KK27 pKa = 10.87KK28 pKa = 9.36KK29 pKa = 10.77
MM1 pKa = 7.84PNGKK5 pKa = 8.55KK6 pKa = 9.72HH7 pKa = 6.1KK8 pKa = 7.52RR9 pKa = 11.84HH10 pKa = 5.93KK11 pKa = 9.95MATHH15 pKa = 6.02KK16 pKa = 10.34RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.26NRR25 pKa = 11.84HH26 pKa = 4.71KK27 pKa = 10.87KK28 pKa = 9.36KK29 pKa = 10.77
Molecular weight: 3.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
781890 |
29 |
1639 |
355.4 |
39.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.469 ± 0.063 | 1.34 ± 0.02 |
5.699 ± 0.04 | 6.722 ± 0.047 |
4.161 ± 0.032 | 7.835 ± 0.047 |
1.921 ± 0.028 | 5.556 ± 0.039 |
3.889 ± 0.054 | 9.385 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.517 ± 0.023 | 3.547 ± 0.045 |
4.333 ± 0.032 | 3.008 ± 0.029 |
7.226 ± 0.064 | 5.655 ± 0.041 |
5.54 ± 0.041 | 7.078 ± 0.05 |
1.293 ± 0.024 | 3.822 ± 0.044 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
