
White sucker hepatitis B virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; Parahepadnavirus
Average proteome isoelectric point is 9.0
Get precalculated fractions of proteins
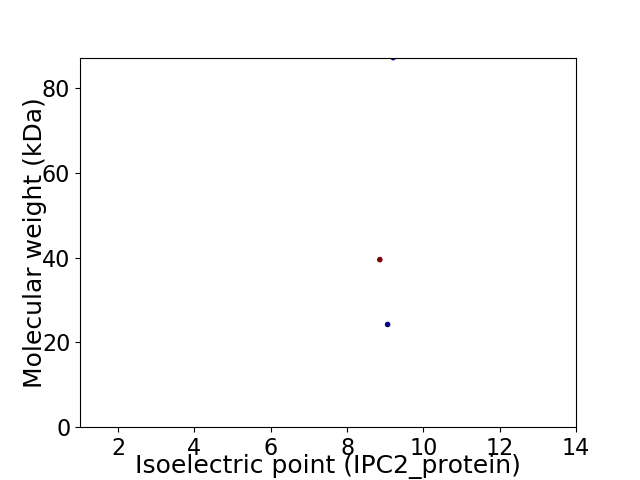
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
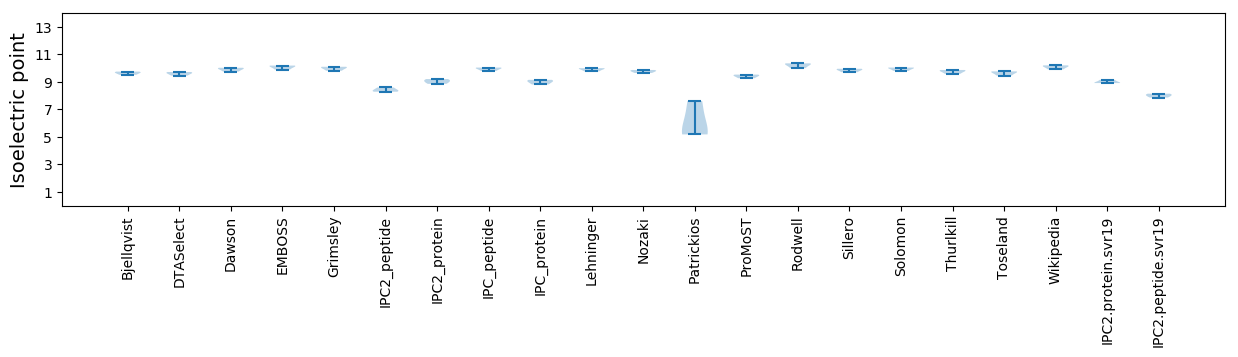
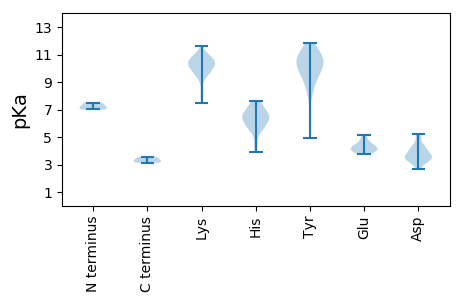
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M3YV36|A0A0M3YV36_9HEPA DNA-directed DNA polymerase OS=White sucker hepatitis B virus OX=1690672 GN=wsHBV_gp2 PE=3 SV=1
MM1 pKa = 7.51GKK3 pKa = 10.41GSSKK7 pKa = 10.95LMTYY11 pKa = 10.57EE12 pKa = 3.88EE13 pKa = 4.51AQHH16 pKa = 4.73QRR18 pKa = 11.84RR19 pKa = 11.84MEE21 pKa = 3.88QYY23 pKa = 8.65MDD25 pKa = 3.41QQARR29 pKa = 11.84TALQRR34 pKa = 11.84LVSQSPARR42 pKa = 11.84DD43 pKa = 3.52VNKK46 pKa = 9.81TKK48 pKa = 10.72SLQWPKK54 pKa = 10.91LPDD57 pKa = 3.62PVTIHH62 pKa = 6.38KK63 pKa = 10.02PSYY66 pKa = 9.55QGSVPPKK73 pKa = 9.87RR74 pKa = 11.84PRR76 pKa = 11.84TEE78 pKa = 3.88LDD80 pKa = 3.0QPLDD84 pKa = 3.78HH85 pKa = 6.29QQSVPLQAPQTVLQPLEE102 pKa = 4.07PPPRR106 pKa = 11.84VMQIEE111 pKa = 4.44QPLQSPPLASKK122 pKa = 10.38VSPLSPIKK130 pKa = 10.27AVPFLAPLLIPVAHH144 pKa = 4.66QTVRR148 pKa = 11.84TSYY151 pKa = 11.83NMTTSNISEE160 pKa = 4.16IMDD163 pKa = 3.7EE164 pKa = 4.09ARR166 pKa = 11.84TAFTTMGSAFQGLSFDD182 pKa = 3.41LWVLFLVVLLVVFFLLIKK200 pKa = 10.17IHH202 pKa = 6.74TILKK206 pKa = 9.56AADD209 pKa = 2.67WWLISRR215 pKa = 11.84SFLGTQTCPFRR226 pKa = 11.84NTEE229 pKa = 4.43SPTSMHH235 pKa = 6.87FKK237 pKa = 9.69TDD239 pKa = 3.37CPLTCTGFRR248 pKa = 11.84WTLVRR253 pKa = 11.84RR254 pKa = 11.84SIIFLCILALVVIFWYY270 pKa = 10.67LMAWEE275 pKa = 4.92PFTAFVKK282 pKa = 10.61RR283 pKa = 11.84LWEE286 pKa = 4.42LGSVLGSYY294 pKa = 9.88IFSHH298 pKa = 7.04LSSQLGSVPTIQFIALLTWMIFYY321 pKa = 10.53SHH323 pKa = 6.72IPVWLHH329 pKa = 5.62FLLSFVTSWHH339 pKa = 5.96FLQNGGFF346 pKa = 3.55
MM1 pKa = 7.51GKK3 pKa = 10.41GSSKK7 pKa = 10.95LMTYY11 pKa = 10.57EE12 pKa = 3.88EE13 pKa = 4.51AQHH16 pKa = 4.73QRR18 pKa = 11.84RR19 pKa = 11.84MEE21 pKa = 3.88QYY23 pKa = 8.65MDD25 pKa = 3.41QQARR29 pKa = 11.84TALQRR34 pKa = 11.84LVSQSPARR42 pKa = 11.84DD43 pKa = 3.52VNKK46 pKa = 9.81TKK48 pKa = 10.72SLQWPKK54 pKa = 10.91LPDD57 pKa = 3.62PVTIHH62 pKa = 6.38KK63 pKa = 10.02PSYY66 pKa = 9.55QGSVPPKK73 pKa = 9.87RR74 pKa = 11.84PRR76 pKa = 11.84TEE78 pKa = 3.88LDD80 pKa = 3.0QPLDD84 pKa = 3.78HH85 pKa = 6.29QQSVPLQAPQTVLQPLEE102 pKa = 4.07PPPRR106 pKa = 11.84VMQIEE111 pKa = 4.44QPLQSPPLASKK122 pKa = 10.38VSPLSPIKK130 pKa = 10.27AVPFLAPLLIPVAHH144 pKa = 4.66QTVRR148 pKa = 11.84TSYY151 pKa = 11.83NMTTSNISEE160 pKa = 4.16IMDD163 pKa = 3.7EE164 pKa = 4.09ARR166 pKa = 11.84TAFTTMGSAFQGLSFDD182 pKa = 3.41LWVLFLVVLLVVFFLLIKK200 pKa = 10.17IHH202 pKa = 6.74TILKK206 pKa = 9.56AADD209 pKa = 2.67WWLISRR215 pKa = 11.84SFLGTQTCPFRR226 pKa = 11.84NTEE229 pKa = 4.43SPTSMHH235 pKa = 6.87FKK237 pKa = 9.69TDD239 pKa = 3.37CPLTCTGFRR248 pKa = 11.84WTLVRR253 pKa = 11.84RR254 pKa = 11.84SIIFLCILALVVIFWYY270 pKa = 10.67LMAWEE275 pKa = 4.92PFTAFVKK282 pKa = 10.61RR283 pKa = 11.84LWEE286 pKa = 4.42LGSVLGSYY294 pKa = 9.88IFSHH298 pKa = 7.04LSSQLGSVPTIQFIALLTWMIFYY321 pKa = 10.53SHH323 pKa = 6.72IPVWLHH329 pKa = 5.62FLLSFVTSWHH339 pKa = 5.96FLQNGGFF346 pKa = 3.55
Molecular weight: 39.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M3P4Q2|A0A0M3P4Q2_9HEPA Large envelope protein OS=White sucker hepatitis B virus OX=1690672 GN=wsHBV_gp3 PE=4 SV=1
MM1 pKa = 7.16FAFVLILLRR10 pKa = 11.84LWSGCLNGSSTMDD23 pKa = 4.19PLVKK27 pKa = 10.27AALEE31 pKa = 4.26GVKK34 pKa = 10.3DD35 pKa = 3.88LPHH38 pKa = 7.5DD39 pKa = 4.28FFPLLKK45 pKa = 10.77DD46 pKa = 3.26QVQFSKK52 pKa = 11.27DD53 pKa = 3.33VIKK56 pKa = 10.35EE57 pKa = 4.08YY58 pKa = 10.99NEE60 pKa = 3.75HH61 pKa = 6.72HH62 pKa = 5.95SQNRR66 pKa = 11.84HH67 pKa = 4.16VEE69 pKa = 4.36VINCSLEE76 pKa = 4.53SYY78 pKa = 11.25DD79 pKa = 4.39NLQLIYY85 pKa = 10.98AKK87 pKa = 9.37VTISNWTAIANNGDD101 pKa = 3.98SVQVALDD108 pKa = 3.52EE109 pKa = 4.17NARR112 pKa = 11.84TGLAYY117 pKa = 9.76IFQSTIEE124 pKa = 3.73MSIRR128 pKa = 11.84RR129 pKa = 11.84KK130 pKa = 9.91LWWHH134 pKa = 5.95TNCLMWGEE142 pKa = 4.41SQTAEE147 pKa = 4.02YY148 pKa = 8.24TAKK151 pKa = 10.31LRR153 pKa = 11.84SWLMTPTSYY162 pKa = 11.07RR163 pKa = 11.84NQYY166 pKa = 10.6APTIEE171 pKa = 4.11ALTRR175 pKa = 11.84TVKK178 pKa = 9.51ATHH181 pKa = 6.39LSSRR185 pKa = 11.84SSRR188 pKa = 11.84PTARR192 pKa = 11.84SPGVRR197 pKa = 11.84ARR199 pKa = 11.84SSSKK203 pKa = 10.75SKK205 pKa = 10.64SKK207 pKa = 10.62SPRR210 pKa = 11.84RR211 pKa = 11.84KK212 pKa = 7.49TT213 pKa = 3.29
MM1 pKa = 7.16FAFVLILLRR10 pKa = 11.84LWSGCLNGSSTMDD23 pKa = 4.19PLVKK27 pKa = 10.27AALEE31 pKa = 4.26GVKK34 pKa = 10.3DD35 pKa = 3.88LPHH38 pKa = 7.5DD39 pKa = 4.28FFPLLKK45 pKa = 10.77DD46 pKa = 3.26QVQFSKK52 pKa = 11.27DD53 pKa = 3.33VIKK56 pKa = 10.35EE57 pKa = 4.08YY58 pKa = 10.99NEE60 pKa = 3.75HH61 pKa = 6.72HH62 pKa = 5.95SQNRR66 pKa = 11.84HH67 pKa = 4.16VEE69 pKa = 4.36VINCSLEE76 pKa = 4.53SYY78 pKa = 11.25DD79 pKa = 4.39NLQLIYY85 pKa = 10.98AKK87 pKa = 9.37VTISNWTAIANNGDD101 pKa = 3.98SVQVALDD108 pKa = 3.52EE109 pKa = 4.17NARR112 pKa = 11.84TGLAYY117 pKa = 9.76IFQSTIEE124 pKa = 3.73MSIRR128 pKa = 11.84RR129 pKa = 11.84KK130 pKa = 9.91LWWHH134 pKa = 5.95TNCLMWGEE142 pKa = 4.41SQTAEE147 pKa = 4.02YY148 pKa = 8.24TAKK151 pKa = 10.31LRR153 pKa = 11.84SWLMTPTSYY162 pKa = 11.07RR163 pKa = 11.84NQYY166 pKa = 10.6APTIEE171 pKa = 4.11ALTRR175 pKa = 11.84TVKK178 pKa = 9.51ATHH181 pKa = 6.39LSSRR185 pKa = 11.84SSRR188 pKa = 11.84PTARR192 pKa = 11.84SPGVRR197 pKa = 11.84ARR199 pKa = 11.84SSSKK203 pKa = 10.75SKK205 pKa = 10.64SKK207 pKa = 10.62SPRR210 pKa = 11.84RR211 pKa = 11.84KK212 pKa = 7.49TT213 pKa = 3.29
Molecular weight: 24.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1347 |
213 |
788 |
449.0 |
50.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.35 ± 0.891 | 1.633 ± 0.236 |
3.267 ± 0.24 | 2.747 ± 0.584 |
4.826 ± 0.559 | 4.232 ± 0.485 |
3.638 ± 0.454 | 4.9 ± 0.228 |
5.048 ± 0.462 | 11.359 ± 0.485 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.856 ± 0.633 | 3.192 ± 0.625 |
7.275 ± 0.702 | 4.677 ± 0.86 |
5.271 ± 0.356 | 10.69 ± 0.635 |
6.904 ± 0.359 | 5.939 ± 0.389 |
2.153 ± 0.538 | 3.044 ± 0.405 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
