
Hubei polero-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
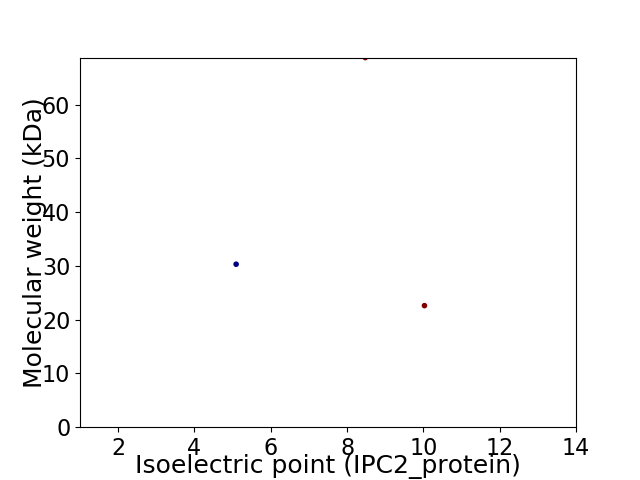
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KF08|A0A1L3KF08_9VIRU Serine protease OS=Hubei polero-like virus 1 OX=1923169 PE=4 SV=1
MM1 pKa = 7.27SVSLVDD7 pKa = 3.44QLVARR12 pKa = 11.84VLFQNQNKK20 pKa = 9.66RR21 pKa = 11.84EE22 pKa = 3.94IALWRR27 pKa = 11.84ALPSKK32 pKa = 10.37PGFGLSTDD40 pKa = 3.67GQVLDD45 pKa = 5.29FVQCLARR52 pKa = 11.84QINVPSEE59 pKa = 3.96EE60 pKa = 4.05VVEE63 pKa = 3.93NWEE66 pKa = 4.35RR67 pKa = 11.84YY68 pKa = 8.75LVPTDD73 pKa = 3.61CSGFDD78 pKa = 3.21WSVAEE83 pKa = 5.47WMLQDD88 pKa = 4.67DD89 pKa = 4.11MEE91 pKa = 4.38VRR93 pKa = 11.84NRR95 pKa = 11.84LTTDD99 pKa = 4.09LNPLTRR105 pKa = 11.84RR106 pKa = 11.84LRR108 pKa = 11.84AGWLKK113 pKa = 10.66CLSNSVLSLSDD124 pKa = 3.28GTLLAQRR131 pKa = 11.84VPGVQKK137 pKa = 10.45SGSYY141 pKa = 8.27NTSSTNSRR149 pKa = 11.84IRR151 pKa = 11.84VMAAYY156 pKa = 9.95HH157 pKa = 6.63CGADD161 pKa = 3.2WAIAMGDD168 pKa = 3.79DD169 pKa = 3.96ALEE172 pKa = 4.34SANSNLEE179 pKa = 4.08EE180 pKa = 4.12YY181 pKa = 10.45KK182 pKa = 10.88SLGFKK187 pKa = 10.75VEE189 pKa = 3.93VSGQLEE195 pKa = 4.67FCSHH199 pKa = 5.47TFVRR203 pKa = 11.84PDD205 pKa = 3.0LALPVNIGKK214 pKa = 8.35MLYY217 pKa = 10.02KK218 pKa = 10.68LIYY221 pKa = 9.82GYY223 pKa = 10.31EE224 pKa = 4.1AGSGSLEE231 pKa = 4.17VVTNYY236 pKa = 10.7LNACFAVLNEE246 pKa = 4.34LRR248 pKa = 11.84HH249 pKa = 5.99DD250 pKa = 4.15PEE252 pKa = 4.61TVRR255 pKa = 11.84LLYY258 pKa = 9.99QWLVLPVQPQNEE270 pKa = 4.13
MM1 pKa = 7.27SVSLVDD7 pKa = 3.44QLVARR12 pKa = 11.84VLFQNQNKK20 pKa = 9.66RR21 pKa = 11.84EE22 pKa = 3.94IALWRR27 pKa = 11.84ALPSKK32 pKa = 10.37PGFGLSTDD40 pKa = 3.67GQVLDD45 pKa = 5.29FVQCLARR52 pKa = 11.84QINVPSEE59 pKa = 3.96EE60 pKa = 4.05VVEE63 pKa = 3.93NWEE66 pKa = 4.35RR67 pKa = 11.84YY68 pKa = 8.75LVPTDD73 pKa = 3.61CSGFDD78 pKa = 3.21WSVAEE83 pKa = 5.47WMLQDD88 pKa = 4.67DD89 pKa = 4.11MEE91 pKa = 4.38VRR93 pKa = 11.84NRR95 pKa = 11.84LTTDD99 pKa = 4.09LNPLTRR105 pKa = 11.84RR106 pKa = 11.84LRR108 pKa = 11.84AGWLKK113 pKa = 10.66CLSNSVLSLSDD124 pKa = 3.28GTLLAQRR131 pKa = 11.84VPGVQKK137 pKa = 10.45SGSYY141 pKa = 8.27NTSSTNSRR149 pKa = 11.84IRR151 pKa = 11.84VMAAYY156 pKa = 9.95HH157 pKa = 6.63CGADD161 pKa = 3.2WAIAMGDD168 pKa = 3.79DD169 pKa = 3.96ALEE172 pKa = 4.34SANSNLEE179 pKa = 4.08EE180 pKa = 4.12YY181 pKa = 10.45KK182 pKa = 10.88SLGFKK187 pKa = 10.75VEE189 pKa = 3.93VSGQLEE195 pKa = 4.67FCSHH199 pKa = 5.47TFVRR203 pKa = 11.84PDD205 pKa = 3.0LALPVNIGKK214 pKa = 8.35MLYY217 pKa = 10.02KK218 pKa = 10.68LIYY221 pKa = 9.82GYY223 pKa = 10.31EE224 pKa = 4.1AGSGSLEE231 pKa = 4.17VVTNYY236 pKa = 10.7LNACFAVLNEE246 pKa = 4.34LRR248 pKa = 11.84HH249 pKa = 5.99DD250 pKa = 4.15PEE252 pKa = 4.61TVRR255 pKa = 11.84LLYY258 pKa = 9.99QWLVLPVQPQNEE270 pKa = 4.13
Molecular weight: 30.31 kDa
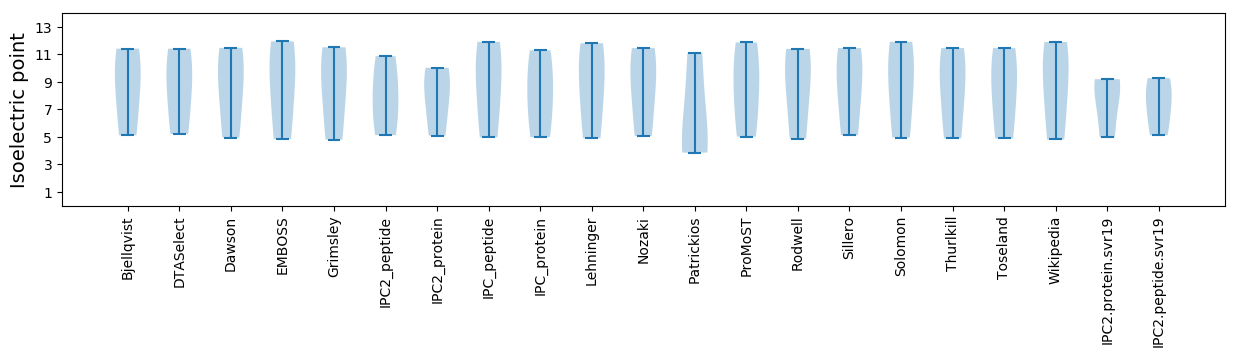
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF55|A0A1L3KF55_9VIRU Uncharacterized protein OS=Hubei polero-like virus 1 OX=1923169 PE=4 SV=1
MM1 pKa = 6.52NTVVVRR7 pKa = 11.84RR8 pKa = 11.84NGRR11 pKa = 11.84RR12 pKa = 11.84GLRR15 pKa = 11.84LRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84NARR25 pKa = 11.84TQQMVVVTQPQRR37 pKa = 11.84TQRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84QRR45 pKa = 11.84RR46 pKa = 11.84GRR48 pKa = 11.84GTSSRR53 pKa = 11.84RR54 pKa = 11.84GAVGRR59 pKa = 11.84GNNFEE64 pKa = 4.28TFVFSKK70 pKa = 11.1DD71 pKa = 3.43NLKK74 pKa = 10.86GSSKK78 pKa = 9.92GTFTFGPSLSDD89 pKa = 3.45CPAFASGILKK99 pKa = 10.22AYY101 pKa = 9.97HH102 pKa = 6.43EE103 pKa = 4.51YY104 pKa = 10.82KK105 pKa = 10.39IISCNMEE112 pKa = 4.67FISEE116 pKa = 4.23ASSTAAGSIAYY127 pKa = 9.34EE128 pKa = 4.13LDD130 pKa = 3.49PHH132 pKa = 7.36CKK134 pKa = 9.33LTDD137 pKa = 3.64LASTVNKK144 pKa = 10.02FSITKK149 pKa = 9.78GGKK152 pKa = 8.06KK153 pKa = 8.45TFTARR158 pKa = 11.84LINGVEE164 pKa = 3.8WHH166 pKa = 7.3DD167 pKa = 3.52ATEE170 pKa = 4.15DD171 pKa = 3.45QFRR174 pKa = 11.84VHH176 pKa = 6.3YY177 pKa = 9.76KK178 pKa = 11.03GNGDD182 pKa = 3.54STTAGSFRR190 pKa = 11.84ITIRR194 pKa = 11.84VATQNPKK201 pKa = 10.5
MM1 pKa = 6.52NTVVVRR7 pKa = 11.84RR8 pKa = 11.84NGRR11 pKa = 11.84RR12 pKa = 11.84GLRR15 pKa = 11.84LRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84NARR25 pKa = 11.84TQQMVVVTQPQRR37 pKa = 11.84TQRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84QRR45 pKa = 11.84RR46 pKa = 11.84GRR48 pKa = 11.84GTSSRR53 pKa = 11.84RR54 pKa = 11.84GAVGRR59 pKa = 11.84GNNFEE64 pKa = 4.28TFVFSKK70 pKa = 11.1DD71 pKa = 3.43NLKK74 pKa = 10.86GSSKK78 pKa = 9.92GTFTFGPSLSDD89 pKa = 3.45CPAFASGILKK99 pKa = 10.22AYY101 pKa = 9.97HH102 pKa = 6.43EE103 pKa = 4.51YY104 pKa = 10.82KK105 pKa = 10.39IISCNMEE112 pKa = 4.67FISEE116 pKa = 4.23ASSTAAGSIAYY127 pKa = 9.34EE128 pKa = 4.13LDD130 pKa = 3.49PHH132 pKa = 7.36CKK134 pKa = 9.33LTDD137 pKa = 3.64LASTVNKK144 pKa = 10.02FSITKK149 pKa = 9.78GGKK152 pKa = 8.06KK153 pKa = 8.45TFTARR158 pKa = 11.84LINGVEE164 pKa = 3.8WHH166 pKa = 7.3DD167 pKa = 3.52ATEE170 pKa = 4.15DD171 pKa = 3.45QFRR174 pKa = 11.84VHH176 pKa = 6.3YY177 pKa = 9.76KK178 pKa = 11.03GNGDD182 pKa = 3.54STTAGSFRR190 pKa = 11.84ITIRR194 pKa = 11.84VATQNPKK201 pKa = 10.5
Molecular weight: 22.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1101 |
201 |
630 |
367.0 |
40.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.811 ± 0.59 | 1.544 ± 0.254 |
3.361 ± 0.712 | 5.268 ± 0.587 |
4.178 ± 0.515 | 7.357 ± 0.619 |
1.817 ± 0.256 | 3.724 ± 0.544 |
5.086 ± 0.766 | 9.628 ± 1.808 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.635 ± 0.214 | 4.632 ± 0.641 |
5.54 ± 1.299 | 3.815 ± 0.403 |
6.358 ± 2.27 | 9.446 ± 0.554 |
6.812 ± 1.115 | 7.175 ± 0.889 |
1.998 ± 0.468 | 2.816 ± 0.277 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
