
Thalassomonas sp. M1454
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Colwelliaceae; Thalassomonas; unclassified Thalassomonas
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
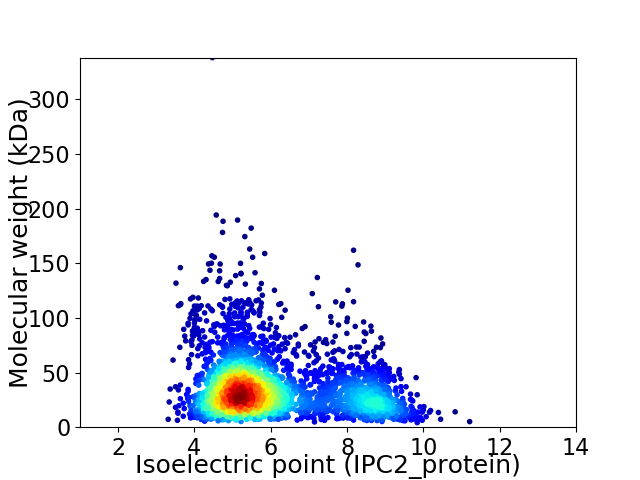
Virtual 2D-PAGE plot for 3137 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A553FLB4|A0A553FLB4_9GAMM DUF416 family protein OS=Thalassomonas sp. M1454 OX=2594477 GN=FNN08_01170 PE=4 SV=1
MM1 pKa = 7.72NSLLKK6 pKa = 9.84KK7 pKa = 7.03TTILAALSVSLLGCDD22 pKa = 5.09DD23 pKa = 3.56VDD25 pKa = 3.78EE26 pKa = 5.87GYY28 pKa = 11.68ANNFKK33 pKa = 10.79NDD35 pKa = 3.43NPVGFSMEE43 pKa = 4.06NIAAEE48 pKa = 4.13FEE50 pKa = 4.47EE51 pKa = 4.72VSGIQQVNLLEE62 pKa = 4.34GAMVDD67 pKa = 4.26GEE69 pKa = 4.47PLTLEE74 pKa = 4.45SSSSPIYY81 pKa = 10.3IRR83 pKa = 11.84DD84 pKa = 3.73FNFEE88 pKa = 4.15TLTDD92 pKa = 3.93GFTTPQAPGFANNQGVSPFTIGDD115 pKa = 3.63DD116 pKa = 3.84GVTLEE121 pKa = 4.52IDD123 pKa = 2.99TDD125 pKa = 3.8AFAEE129 pKa = 4.18ALRR132 pKa = 11.84MCDD135 pKa = 3.27PTDD138 pKa = 3.11VRR140 pKa = 11.84GAVDD144 pKa = 3.79EE145 pKa = 4.32NNNPIGNGVRR155 pKa = 11.84DD156 pKa = 4.29FPTLVEE162 pKa = 3.82YY163 pKa = 10.87SITYY167 pKa = 9.89VVDD170 pKa = 3.03NGYY173 pKa = 10.68EE174 pKa = 3.88YY175 pKa = 11.26APGEE179 pKa = 4.07EE180 pKa = 4.34HH181 pKa = 7.3PVRR184 pKa = 11.84TLEE187 pKa = 3.97LTITAGSDD195 pKa = 3.12PVANVQAFDD204 pKa = 3.76INVASGDD211 pKa = 4.27SVPMLSATAPAYY223 pKa = 10.59SCNSNLTYY231 pKa = 10.83DD232 pKa = 3.68VADD235 pKa = 3.69TSIASVDD242 pKa = 3.61EE243 pKa = 4.45NGVITGNNKK252 pKa = 9.36GQTTITVASVEE263 pKa = 4.24NPEE266 pKa = 4.1LTATATINVTPGFNLLITNQEE287 pKa = 4.18FNDD290 pKa = 3.6LGAPLGTKK298 pKa = 9.83AVPTCSAIGVNVEE311 pKa = 3.78PTIVADD317 pKa = 4.13EE318 pKa = 4.31LSGEE322 pKa = 4.12YY323 pKa = 9.78TYY325 pKa = 11.59AWSSDD330 pKa = 3.4SLVLAKK336 pKa = 10.72EE337 pKa = 4.0EE338 pKa = 4.32TTGGFSATGRR348 pKa = 11.84FANTLAVGEE357 pKa = 4.53SADD360 pKa = 3.28ITVGYY365 pKa = 10.26GSGYY369 pKa = 10.11SGSTAAGDD377 pKa = 3.78VQPQMVTVTAEE388 pKa = 3.95RR389 pKa = 11.84NLACDD394 pKa = 3.93PGLNADD400 pKa = 3.77GTDD403 pKa = 4.39FFASDD408 pKa = 4.28LKK410 pKa = 11.04LDD412 pKa = 3.83SGNWGASATHH422 pKa = 5.58VAEE425 pKa = 4.55GLDD428 pKa = 3.75GTSLQITSTNGGKK441 pKa = 10.31FYY443 pKa = 11.16GGVVQQQWNQVFNYY457 pKa = 9.66HH458 pKa = 6.49AATYY462 pKa = 9.87GRR464 pKa = 11.84GGASIGRR471 pKa = 11.84TFKK474 pKa = 10.86FKK476 pKa = 10.1VWAKK480 pKa = 10.35LGQLPTEE487 pKa = 4.23EE488 pKa = 4.44VKK490 pKa = 10.98LEE492 pKa = 3.89HH493 pKa = 7.39SIIPWNCDD501 pKa = 2.68GCDD504 pKa = 3.49GLANYY509 pKa = 7.89PARR512 pKa = 11.84IEE514 pKa = 4.18AGVAGTVTAVLKK526 pKa = 8.41PTTDD530 pKa = 2.87WQLVEE535 pKa = 5.78FINPLTGTDD544 pKa = 3.29EE545 pKa = 4.33WSVPAHH551 pKa = 5.88FNVSTAAFLSWDD563 pKa = 3.08IYY565 pKa = 11.26GFADD569 pKa = 4.59GEE571 pKa = 4.63TILLDD576 pKa = 3.4NYY578 pKa = 10.96SIVEE582 pKa = 4.05VEE584 pKa = 3.87
MM1 pKa = 7.72NSLLKK6 pKa = 9.84KK7 pKa = 7.03TTILAALSVSLLGCDD22 pKa = 5.09DD23 pKa = 3.56VDD25 pKa = 3.78EE26 pKa = 5.87GYY28 pKa = 11.68ANNFKK33 pKa = 10.79NDD35 pKa = 3.43NPVGFSMEE43 pKa = 4.06NIAAEE48 pKa = 4.13FEE50 pKa = 4.47EE51 pKa = 4.72VSGIQQVNLLEE62 pKa = 4.34GAMVDD67 pKa = 4.26GEE69 pKa = 4.47PLTLEE74 pKa = 4.45SSSSPIYY81 pKa = 10.3IRR83 pKa = 11.84DD84 pKa = 3.73FNFEE88 pKa = 4.15TLTDD92 pKa = 3.93GFTTPQAPGFANNQGVSPFTIGDD115 pKa = 3.63DD116 pKa = 3.84GVTLEE121 pKa = 4.52IDD123 pKa = 2.99TDD125 pKa = 3.8AFAEE129 pKa = 4.18ALRR132 pKa = 11.84MCDD135 pKa = 3.27PTDD138 pKa = 3.11VRR140 pKa = 11.84GAVDD144 pKa = 3.79EE145 pKa = 4.32NNNPIGNGVRR155 pKa = 11.84DD156 pKa = 4.29FPTLVEE162 pKa = 3.82YY163 pKa = 10.87SITYY167 pKa = 9.89VVDD170 pKa = 3.03NGYY173 pKa = 10.68EE174 pKa = 3.88YY175 pKa = 11.26APGEE179 pKa = 4.07EE180 pKa = 4.34HH181 pKa = 7.3PVRR184 pKa = 11.84TLEE187 pKa = 3.97LTITAGSDD195 pKa = 3.12PVANVQAFDD204 pKa = 3.76INVASGDD211 pKa = 4.27SVPMLSATAPAYY223 pKa = 10.59SCNSNLTYY231 pKa = 10.83DD232 pKa = 3.68VADD235 pKa = 3.69TSIASVDD242 pKa = 3.61EE243 pKa = 4.45NGVITGNNKK252 pKa = 9.36GQTTITVASVEE263 pKa = 4.24NPEE266 pKa = 4.1LTATATINVTPGFNLLITNQEE287 pKa = 4.18FNDD290 pKa = 3.6LGAPLGTKK298 pKa = 9.83AVPTCSAIGVNVEE311 pKa = 3.78PTIVADD317 pKa = 4.13EE318 pKa = 4.31LSGEE322 pKa = 4.12YY323 pKa = 9.78TYY325 pKa = 11.59AWSSDD330 pKa = 3.4SLVLAKK336 pKa = 10.72EE337 pKa = 4.0EE338 pKa = 4.32TTGGFSATGRR348 pKa = 11.84FANTLAVGEE357 pKa = 4.53SADD360 pKa = 3.28ITVGYY365 pKa = 10.26GSGYY369 pKa = 10.11SGSTAAGDD377 pKa = 3.78VQPQMVTVTAEE388 pKa = 3.95RR389 pKa = 11.84NLACDD394 pKa = 3.93PGLNADD400 pKa = 3.77GTDD403 pKa = 4.39FFASDD408 pKa = 4.28LKK410 pKa = 11.04LDD412 pKa = 3.83SGNWGASATHH422 pKa = 5.58VAEE425 pKa = 4.55GLDD428 pKa = 3.75GTSLQITSTNGGKK441 pKa = 10.31FYY443 pKa = 11.16GGVVQQQWNQVFNYY457 pKa = 9.66HH458 pKa = 6.49AATYY462 pKa = 9.87GRR464 pKa = 11.84GGASIGRR471 pKa = 11.84TFKK474 pKa = 10.86FKK476 pKa = 10.1VWAKK480 pKa = 10.35LGQLPTEE487 pKa = 4.23EE488 pKa = 4.44VKK490 pKa = 10.98LEE492 pKa = 3.89HH493 pKa = 7.39SIIPWNCDD501 pKa = 2.68GCDD504 pKa = 3.49GLANYY509 pKa = 7.89PARR512 pKa = 11.84IEE514 pKa = 4.18AGVAGTVTAVLKK526 pKa = 8.41PTTDD530 pKa = 2.87WQLVEE535 pKa = 5.78FINPLTGTDD544 pKa = 3.29EE545 pKa = 4.33WSVPAHH551 pKa = 5.88FNVSTAAFLSWDD563 pKa = 3.08IYY565 pKa = 11.26GFADD569 pKa = 4.59GEE571 pKa = 4.63TILLDD576 pKa = 3.4NYY578 pKa = 10.96SIVEE582 pKa = 4.05VEE584 pKa = 3.87
Molecular weight: 61.72 kDa
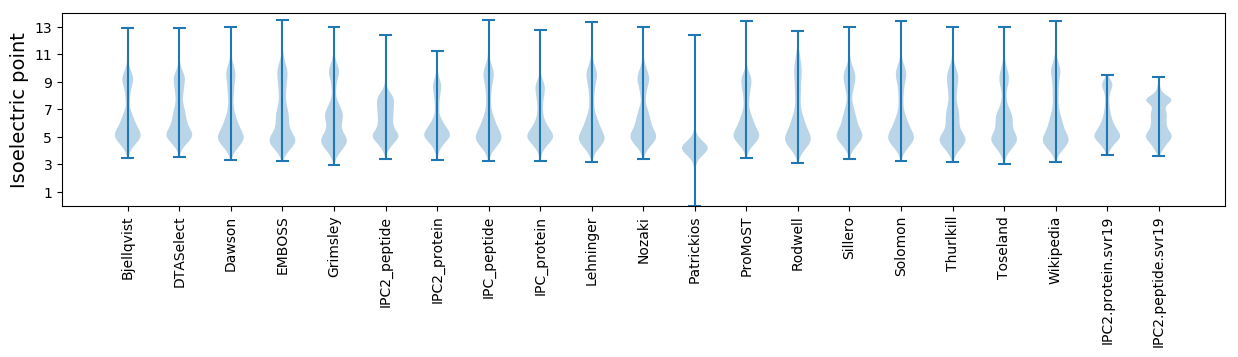
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A553FLL8|A0A553FLL8_9GAMM Probable chromosome-partitioning protein ParB OS=Thalassomonas sp. M1454 OX=2594477 GN=FNN08_01690 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.16RR12 pKa = 11.84KK13 pKa = 9.08RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.33NGRR28 pKa = 11.84AVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.45GRR39 pKa = 11.84AKK41 pKa = 9.64LTAA44 pKa = 4.21
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.16RR12 pKa = 11.84KK13 pKa = 9.08RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.33NGRR28 pKa = 11.84AVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.45GRR39 pKa = 11.84AKK41 pKa = 9.64LTAA44 pKa = 4.21
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1074329 |
37 |
3053 |
342.5 |
38.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.278 ± 0.044 | 0.992 ± 0.015 |
5.805 ± 0.041 | 6.155 ± 0.037 |
4.419 ± 0.031 | 6.556 ± 0.041 |
2.045 ± 0.019 | 7.028 ± 0.034 |
6.067 ± 0.045 | 9.933 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.402 ± 0.022 | 5.17 ± 0.038 |
3.672 ± 0.025 | 4.399 ± 0.035 |
3.7 ± 0.03 | 6.781 ± 0.035 |
5.453 ± 0.03 | 6.778 ± 0.037 |
1.152 ± 0.015 | 3.216 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
