
Micromonospora sp. HK10
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micromonosporales; Micromonosporaceae; Micromonospora; unclassified Micromonospora
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
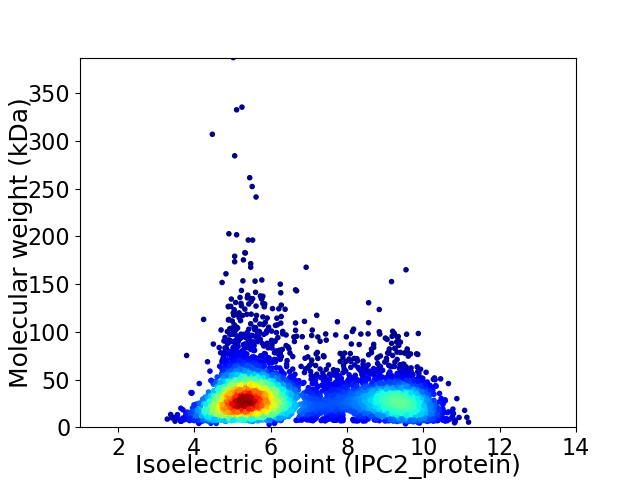
Virtual 2D-PAGE plot for 5548 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M2RH07|A0A0M2RH07_9ACTN MFS transporter OS=Micromonospora sp. HK10 OX=1538294 GN=LQ51_31065 PE=4 SV=1
MM1 pKa = 6.97TALAFTVSAPAQANDD16 pKa = 3.59HH17 pKa = 6.22EE18 pKa = 5.26NIPGEE23 pKa = 4.01PTITVPINDD32 pKa = 3.64SAAGGTYY39 pKa = 10.09NQDD42 pKa = 3.11FTRR45 pKa = 11.84VYY47 pKa = 11.41ANTTPDD53 pKa = 3.31GTPVYY58 pKa = 10.08IYY60 pKa = 10.52VPKK63 pKa = 9.73GTPLDD68 pKa = 3.74GTAPTGVASLDD79 pKa = 3.45PQFDD83 pKa = 4.21HH84 pKa = 7.16NPGDD88 pKa = 3.72EE89 pKa = 4.42FVPCATATAAQFTLTQAQITYY110 pKa = 10.67VGDD113 pKa = 3.35KK114 pKa = 10.18LANQIVAVDD123 pKa = 4.02EE124 pKa = 4.38EE125 pKa = 5.08HH126 pKa = 6.85FGPMDD131 pKa = 3.96AADD134 pKa = 4.22PADD137 pKa = 4.08PASDD141 pKa = 3.6SLVMLVYY148 pKa = 10.79NIQDD152 pKa = 3.32DD153 pKa = 4.43AYY155 pKa = 9.35YY156 pKa = 10.54DD157 pKa = 3.97CAVTSYY163 pKa = 9.73TAGYY167 pKa = 7.63FAPDD171 pKa = 4.87FIDD174 pKa = 4.12SIGMNVISVDD184 pKa = 3.38GLDD187 pKa = 3.07WANRR191 pKa = 11.84VGPNDD196 pKa = 4.05SPWRR200 pKa = 11.84DD201 pKa = 3.06ADD203 pKa = 3.81PANDD207 pKa = 3.72RR208 pKa = 11.84PEE210 pKa = 4.78LYY212 pKa = 9.95EE213 pKa = 3.8GTVAHH218 pKa = 6.3EE219 pKa = 4.42LEE221 pKa = 4.91HH222 pKa = 6.83LLHH225 pKa = 6.67NYY227 pKa = 9.83SDD229 pKa = 4.0PGEE232 pKa = 4.2LSWVDD237 pKa = 3.2EE238 pKa = 4.25GLADD242 pKa = 4.89FAIFLNGLDD251 pKa = 3.84AGGSHH256 pKa = 6.51LTNHH260 pKa = 4.98QVLYY264 pKa = 10.66EE265 pKa = 4.13EE266 pKa = 4.79TSLTRR271 pKa = 11.84WGGSLANYY279 pKa = 8.39GAAFTYY285 pKa = 9.16FQYY288 pKa = 10.98LWEE291 pKa = 4.13QAGGNGDD298 pKa = 4.05GTWTPDD304 pKa = 2.86KK305 pKa = 11.09QYY307 pKa = 11.18DD308 pKa = 3.87GKK310 pKa = 11.05AGDD313 pKa = 4.72LLIKK317 pKa = 9.82TIFEE321 pKa = 4.07EE322 pKa = 4.52QADD325 pKa = 3.96GMTGVQNAIDD335 pKa = 4.61DD336 pKa = 4.3FNAATGANLRR346 pKa = 11.84SAADD350 pKa = 3.93LFRR353 pKa = 11.84DD354 pKa = 3.46WSVAVYY360 pKa = 10.7LDD362 pKa = 5.32DD363 pKa = 4.81EE364 pKa = 5.26DD365 pKa = 5.57SPIYY369 pKa = 9.82DD370 pKa = 2.86IKK372 pKa = 11.21AFNFGDD378 pKa = 3.86PADD381 pKa = 4.44TSWTMDD387 pKa = 3.31IADD390 pKa = 3.65DD391 pKa = 4.46VYY393 pKa = 9.68WGGRR397 pKa = 11.84GKK399 pKa = 10.62YY400 pKa = 9.76HH401 pKa = 7.13GATPEE406 pKa = 4.11AKK408 pKa = 9.21WARR411 pKa = 11.84LKK413 pKa = 10.72NRR415 pKa = 11.84PDD417 pKa = 3.11TTAVPFGMSVEE428 pKa = 4.27RR429 pKa = 11.84FRR431 pKa = 11.84NPGPTVSVAFDD442 pKa = 3.9GDD444 pKa = 4.11DD445 pKa = 3.51EE446 pKa = 5.01TVVPPHH452 pKa = 6.56TGATHH457 pKa = 6.73WYY459 pKa = 9.47GGYY462 pKa = 10.23EE463 pKa = 4.08SQDD466 pKa = 3.59DD467 pKa = 4.51NILDD471 pKa = 3.87VTTSGAVTSLDD482 pKa = 2.99FWSWYY487 pKa = 10.04FIEE490 pKa = 4.87EE491 pKa = 4.05GWDD494 pKa = 3.46FGFVEE499 pKa = 4.83ALVGGNWVTVPLRR512 pKa = 11.84TDD514 pKa = 3.44DD515 pKa = 4.1GQVVTTNDD523 pKa = 3.8DD524 pKa = 3.28PHH526 pKa = 8.04GNNTEE531 pKa = 4.32GNGLTGTSGGEE542 pKa = 4.02YY543 pKa = 10.35FVDD546 pKa = 3.8DD547 pKa = 4.4PVYY550 pKa = 10.6QHH552 pKa = 6.59LTADD556 pKa = 4.52LPAGTTDD563 pKa = 2.73VRR565 pKa = 11.84FRR567 pKa = 11.84YY568 pKa = 8.57STDD571 pKa = 2.88AAYY574 pKa = 9.84MDD576 pKa = 4.32TGWFVDD582 pKa = 3.81DD583 pKa = 4.01VRR585 pKa = 11.84VNGADD590 pKa = 3.38AQLSSAEE597 pKa = 4.38GNWFATDD604 pKa = 4.12GAQDD608 pKa = 3.87NNWTVQVVSHH618 pKa = 7.17CDD620 pKa = 3.26LTPGVTSPGEE630 pKa = 4.28TTDD633 pKa = 4.23GAGNWVYY640 pKa = 10.96RR641 pKa = 11.84STGDD645 pKa = 3.26QFTMSGLRR653 pKa = 11.84TKK655 pKa = 10.3CANGNQDD662 pKa = 4.03DD663 pKa = 4.16FAVIINNLPTGDD675 pKa = 4.23LTYY678 pKa = 11.12LDD680 pKa = 3.93ADD682 pKa = 3.59YY683 pKa = 10.57TFRR686 pKa = 11.84VKK688 pKa = 9.97NTGNAKK694 pKa = 10.2
MM1 pKa = 6.97TALAFTVSAPAQANDD16 pKa = 3.59HH17 pKa = 6.22EE18 pKa = 5.26NIPGEE23 pKa = 4.01PTITVPINDD32 pKa = 3.64SAAGGTYY39 pKa = 10.09NQDD42 pKa = 3.11FTRR45 pKa = 11.84VYY47 pKa = 11.41ANTTPDD53 pKa = 3.31GTPVYY58 pKa = 10.08IYY60 pKa = 10.52VPKK63 pKa = 9.73GTPLDD68 pKa = 3.74GTAPTGVASLDD79 pKa = 3.45PQFDD83 pKa = 4.21HH84 pKa = 7.16NPGDD88 pKa = 3.72EE89 pKa = 4.42FVPCATATAAQFTLTQAQITYY110 pKa = 10.67VGDD113 pKa = 3.35KK114 pKa = 10.18LANQIVAVDD123 pKa = 4.02EE124 pKa = 4.38EE125 pKa = 5.08HH126 pKa = 6.85FGPMDD131 pKa = 3.96AADD134 pKa = 4.22PADD137 pKa = 4.08PASDD141 pKa = 3.6SLVMLVYY148 pKa = 10.79NIQDD152 pKa = 3.32DD153 pKa = 4.43AYY155 pKa = 9.35YY156 pKa = 10.54DD157 pKa = 3.97CAVTSYY163 pKa = 9.73TAGYY167 pKa = 7.63FAPDD171 pKa = 4.87FIDD174 pKa = 4.12SIGMNVISVDD184 pKa = 3.38GLDD187 pKa = 3.07WANRR191 pKa = 11.84VGPNDD196 pKa = 4.05SPWRR200 pKa = 11.84DD201 pKa = 3.06ADD203 pKa = 3.81PANDD207 pKa = 3.72RR208 pKa = 11.84PEE210 pKa = 4.78LYY212 pKa = 9.95EE213 pKa = 3.8GTVAHH218 pKa = 6.3EE219 pKa = 4.42LEE221 pKa = 4.91HH222 pKa = 6.83LLHH225 pKa = 6.67NYY227 pKa = 9.83SDD229 pKa = 4.0PGEE232 pKa = 4.2LSWVDD237 pKa = 3.2EE238 pKa = 4.25GLADD242 pKa = 4.89FAIFLNGLDD251 pKa = 3.84AGGSHH256 pKa = 6.51LTNHH260 pKa = 4.98QVLYY264 pKa = 10.66EE265 pKa = 4.13EE266 pKa = 4.79TSLTRR271 pKa = 11.84WGGSLANYY279 pKa = 8.39GAAFTYY285 pKa = 9.16FQYY288 pKa = 10.98LWEE291 pKa = 4.13QAGGNGDD298 pKa = 4.05GTWTPDD304 pKa = 2.86KK305 pKa = 11.09QYY307 pKa = 11.18DD308 pKa = 3.87GKK310 pKa = 11.05AGDD313 pKa = 4.72LLIKK317 pKa = 9.82TIFEE321 pKa = 4.07EE322 pKa = 4.52QADD325 pKa = 3.96GMTGVQNAIDD335 pKa = 4.61DD336 pKa = 4.3FNAATGANLRR346 pKa = 11.84SAADD350 pKa = 3.93LFRR353 pKa = 11.84DD354 pKa = 3.46WSVAVYY360 pKa = 10.7LDD362 pKa = 5.32DD363 pKa = 4.81EE364 pKa = 5.26DD365 pKa = 5.57SPIYY369 pKa = 9.82DD370 pKa = 2.86IKK372 pKa = 11.21AFNFGDD378 pKa = 3.86PADD381 pKa = 4.44TSWTMDD387 pKa = 3.31IADD390 pKa = 3.65DD391 pKa = 4.46VYY393 pKa = 9.68WGGRR397 pKa = 11.84GKK399 pKa = 10.62YY400 pKa = 9.76HH401 pKa = 7.13GATPEE406 pKa = 4.11AKK408 pKa = 9.21WARR411 pKa = 11.84LKK413 pKa = 10.72NRR415 pKa = 11.84PDD417 pKa = 3.11TTAVPFGMSVEE428 pKa = 4.27RR429 pKa = 11.84FRR431 pKa = 11.84NPGPTVSVAFDD442 pKa = 3.9GDD444 pKa = 4.11DD445 pKa = 3.51EE446 pKa = 5.01TVVPPHH452 pKa = 6.56TGATHH457 pKa = 6.73WYY459 pKa = 9.47GGYY462 pKa = 10.23EE463 pKa = 4.08SQDD466 pKa = 3.59DD467 pKa = 4.51NILDD471 pKa = 3.87VTTSGAVTSLDD482 pKa = 2.99FWSWYY487 pKa = 10.04FIEE490 pKa = 4.87EE491 pKa = 4.05GWDD494 pKa = 3.46FGFVEE499 pKa = 4.83ALVGGNWVTVPLRR512 pKa = 11.84TDD514 pKa = 3.44DD515 pKa = 4.1GQVVTTNDD523 pKa = 3.8DD524 pKa = 3.28PHH526 pKa = 8.04GNNTEE531 pKa = 4.32GNGLTGTSGGEE542 pKa = 4.02YY543 pKa = 10.35FVDD546 pKa = 3.8DD547 pKa = 4.4PVYY550 pKa = 10.6QHH552 pKa = 6.59LTADD556 pKa = 4.52LPAGTTDD563 pKa = 2.73VRR565 pKa = 11.84FRR567 pKa = 11.84YY568 pKa = 8.57STDD571 pKa = 2.88AAYY574 pKa = 9.84MDD576 pKa = 4.32TGWFVDD582 pKa = 3.81DD583 pKa = 4.01VRR585 pKa = 11.84VNGADD590 pKa = 3.38AQLSSAEE597 pKa = 4.38GNWFATDD604 pKa = 4.12GAQDD608 pKa = 3.87NNWTVQVVSHH618 pKa = 7.17CDD620 pKa = 3.26LTPGVTSPGEE630 pKa = 4.28TTDD633 pKa = 4.23GAGNWVYY640 pKa = 10.96RR641 pKa = 11.84STGDD645 pKa = 3.26QFTMSGLRR653 pKa = 11.84TKK655 pKa = 10.3CANGNQDD662 pKa = 4.03DD663 pKa = 4.16FAVIINNLPTGDD675 pKa = 4.23LTYY678 pKa = 11.12LDD680 pKa = 3.93ADD682 pKa = 3.59YY683 pKa = 10.57TFRR686 pKa = 11.84VKK688 pKa = 9.97NTGNAKK694 pKa = 10.2
Molecular weight: 75.21 kDa
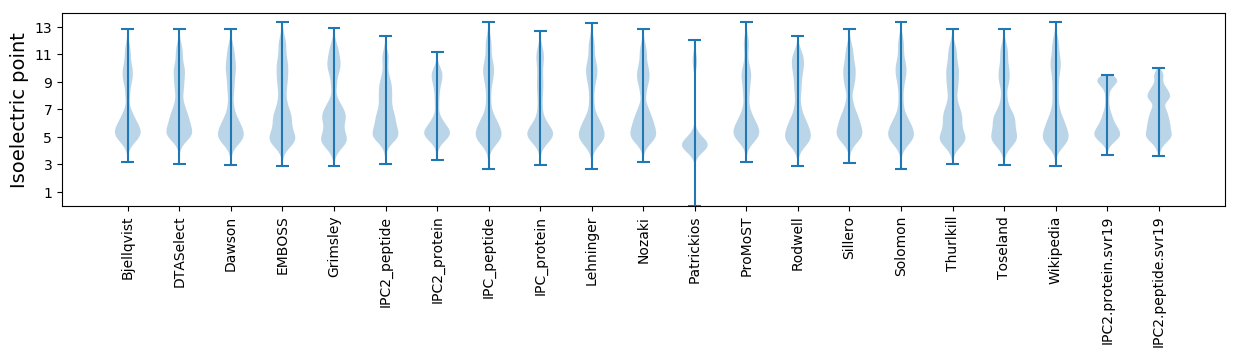
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M2RXK1|A0A0M2RXK1_9ACTN Alanine--tRNA ligase OS=Micromonospora sp. HK10 OX=1538294 GN=alaS PE=3 SV=1
MM1 pKa = 7.71RR2 pKa = 11.84RR3 pKa = 11.84TTTRR7 pKa = 11.84PRR9 pKa = 11.84IRR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84RR15 pKa = 11.84PAGRR19 pKa = 11.84PRR21 pKa = 11.84IRR23 pKa = 11.84RR24 pKa = 11.84LRR26 pKa = 11.84RR27 pKa = 11.84TPTRR31 pKa = 11.84PRR33 pKa = 11.84IRR35 pKa = 11.84RR36 pKa = 11.84LRR38 pKa = 11.84RR39 pKa = 11.84TPTRR43 pKa = 11.84PRR45 pKa = 11.84IRR47 pKa = 11.84RR48 pKa = 11.84LRR50 pKa = 11.84RR51 pKa = 11.84AGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84VRR58 pKa = 11.84RR59 pKa = 11.84AGRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84PARR67 pKa = 11.84RR68 pKa = 11.84GRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84GGRR75 pKa = 11.84PAGRR79 pKa = 11.84RR80 pKa = 11.84VRR82 pKa = 11.84AGWAGGGVGHH92 pKa = 7.09PRR94 pKa = 11.84DD95 pKa = 3.84PRR97 pKa = 11.84AGDD100 pKa = 3.28RR101 pKa = 11.84GAAAPPAGRR110 pKa = 11.84RR111 pKa = 11.84HH112 pKa = 6.07HH113 pKa = 6.94PGAPVRR119 pKa = 11.84CRR121 pKa = 11.84SRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84GGPRR129 pKa = 11.84PAGRR133 pKa = 11.84RR134 pKa = 11.84TPRR137 pKa = 11.84PPPPPCATRR146 pKa = 11.84CPTPAPDD153 pKa = 3.25GG154 pKa = 3.94
MM1 pKa = 7.71RR2 pKa = 11.84RR3 pKa = 11.84TTTRR7 pKa = 11.84PRR9 pKa = 11.84IRR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84RR15 pKa = 11.84PAGRR19 pKa = 11.84PRR21 pKa = 11.84IRR23 pKa = 11.84RR24 pKa = 11.84LRR26 pKa = 11.84RR27 pKa = 11.84TPTRR31 pKa = 11.84PRR33 pKa = 11.84IRR35 pKa = 11.84RR36 pKa = 11.84LRR38 pKa = 11.84RR39 pKa = 11.84TPTRR43 pKa = 11.84PRR45 pKa = 11.84IRR47 pKa = 11.84RR48 pKa = 11.84LRR50 pKa = 11.84RR51 pKa = 11.84AGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84VRR58 pKa = 11.84RR59 pKa = 11.84AGRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84PARR67 pKa = 11.84RR68 pKa = 11.84GRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84GGRR75 pKa = 11.84PAGRR79 pKa = 11.84RR80 pKa = 11.84VRR82 pKa = 11.84AGWAGGGVGHH92 pKa = 7.09PRR94 pKa = 11.84DD95 pKa = 3.84PRR97 pKa = 11.84AGDD100 pKa = 3.28RR101 pKa = 11.84GAAAPPAGRR110 pKa = 11.84RR111 pKa = 11.84HH112 pKa = 6.07HH113 pKa = 6.94PGAPVRR119 pKa = 11.84CRR121 pKa = 11.84SRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84GGPRR129 pKa = 11.84PAGRR133 pKa = 11.84RR134 pKa = 11.84TPRR137 pKa = 11.84PPPPPCATRR146 pKa = 11.84CPTPAPDD153 pKa = 3.25GG154 pKa = 3.94
Molecular weight: 17.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1776529 |
29 |
3736 |
320.2 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.421 ± 0.052 | 0.72 ± 0.01 |
6.047 ± 0.026 | 5.064 ± 0.031 |
2.63 ± 0.02 | 9.5 ± 0.033 |
2.127 ± 0.019 | 3.086 ± 0.022 |
1.609 ± 0.024 | 10.77 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.544 ± 0.011 | 1.724 ± 0.019 |
6.43 ± 0.031 | 2.696 ± 0.019 |
8.583 ± 0.036 | 4.407 ± 0.023 |
5.974 ± 0.031 | 8.99 ± 0.031 |
1.607 ± 0.015 | 2.071 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
