
Adelaide River virus (ARV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ephemerovirus; Adelaide River ephemerovirus
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
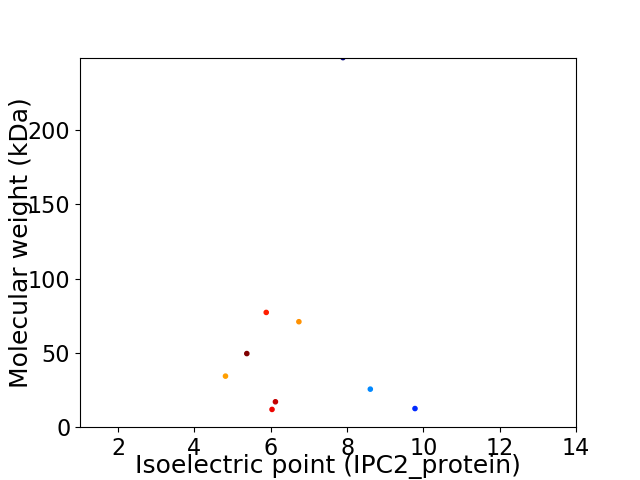
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
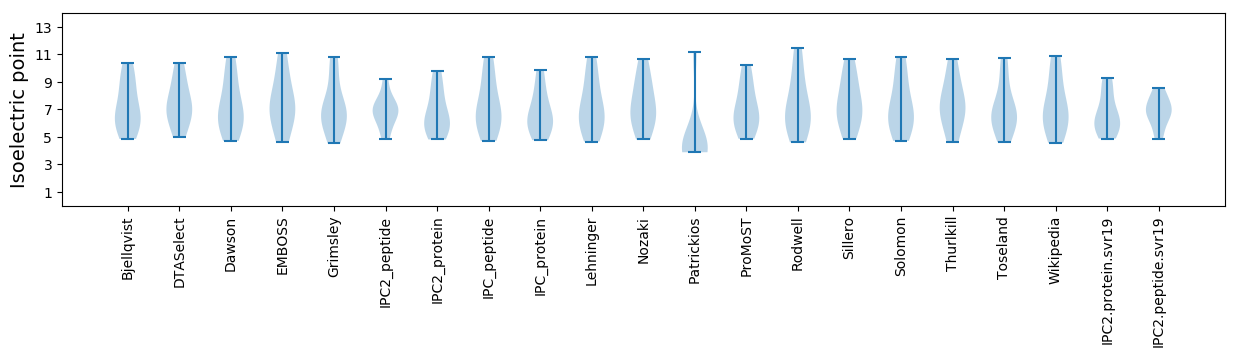
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4HH67|K4HH67_ARV Beta protein OS=Adelaide River virus OX=31612 GN=beta PE=4 SV=1
MM1 pKa = 7.6EE2 pKa = 5.49KK3 pKa = 10.51FNPSKK8 pKa = 11.13VLGEE12 pKa = 4.11YY13 pKa = 9.21DD14 pKa = 3.57TDD16 pKa = 4.18KK17 pKa = 11.38LIASINEE24 pKa = 4.43TNWNLDD30 pKa = 3.41DD31 pKa = 5.58DD32 pKa = 5.12PEE34 pKa = 4.47FSAPQEE40 pKa = 4.31SIEE43 pKa = 4.19PSNSIDD49 pKa = 3.37NYY51 pKa = 9.63RR52 pKa = 11.84DD53 pKa = 3.5EE54 pKa = 4.06IVKK57 pKa = 10.36IGVCLSSNNQDD68 pKa = 3.35YY69 pKa = 11.6SFLDD73 pKa = 3.43NQKK76 pKa = 9.89EE77 pKa = 4.06KK78 pKa = 10.96EE79 pKa = 4.04IFLEE83 pKa = 4.67EE84 pKa = 4.0EE85 pKa = 3.92NDD87 pKa = 3.54WEE89 pKa = 4.85VEE91 pKa = 4.29FTKK94 pKa = 10.74KK95 pKa = 10.84LEE97 pKa = 4.07DD98 pKa = 3.46HH99 pKa = 6.44NVPNEE104 pKa = 4.13SEE106 pKa = 4.89LEE108 pKa = 4.06VQINIEE114 pKa = 4.06DD115 pKa = 5.14LIALLHH121 pKa = 5.76FLNVEE126 pKa = 3.71IDD128 pKa = 3.43QYY130 pKa = 11.91NILPLSLQNKK140 pKa = 9.52VILQRR145 pKa = 11.84KK146 pKa = 9.12KK147 pKa = 10.45EE148 pKa = 4.06GKK150 pKa = 9.84SDD152 pKa = 3.49DD153 pKa = 4.06KK154 pKa = 11.25KK155 pKa = 10.39RR156 pKa = 11.84QNGKK160 pKa = 9.83GDD162 pKa = 3.66KK163 pKa = 10.24KK164 pKa = 10.94SCPDD168 pKa = 4.58SKK170 pKa = 11.17NQSNNCQTMNAEE182 pKa = 4.69TNGSTNLGADD192 pKa = 3.65RR193 pKa = 11.84GPNQTEE199 pKa = 4.57DD200 pKa = 3.44NSSMLEE206 pKa = 3.96NDD208 pKa = 3.4SEE210 pKa = 5.74AILDD214 pKa = 3.76SDD216 pKa = 4.4GGEE219 pKa = 3.79LFQFIIKK226 pKa = 9.65TMDD229 pKa = 2.66QGIRR233 pKa = 11.84VKK235 pKa = 10.8KK236 pKa = 9.93KK237 pKa = 9.66FSGKK241 pKa = 8.99KK242 pKa = 9.31VKK244 pKa = 8.69ITRR247 pKa = 11.84EE248 pKa = 4.08NIGLDD253 pKa = 3.08YY254 pKa = 11.02HH255 pKa = 6.95VINGHH260 pKa = 4.91IHH262 pKa = 6.03GKK264 pKa = 7.33SAKK267 pKa = 10.29DD268 pKa = 3.52SIDD271 pKa = 3.28LKK273 pKa = 11.19KK274 pKa = 10.92LMRR277 pKa = 11.84EE278 pKa = 4.37LIKK281 pKa = 10.61KK282 pKa = 7.77GRR284 pKa = 11.84KK285 pKa = 8.52YY286 pKa = 10.86KK287 pKa = 10.57DD288 pKa = 3.21YY289 pKa = 11.42SNQLDD294 pKa = 3.51LSEE297 pKa = 5.02IEE299 pKa = 4.29FF300 pKa = 4.22
MM1 pKa = 7.6EE2 pKa = 5.49KK3 pKa = 10.51FNPSKK8 pKa = 11.13VLGEE12 pKa = 4.11YY13 pKa = 9.21DD14 pKa = 3.57TDD16 pKa = 4.18KK17 pKa = 11.38LIASINEE24 pKa = 4.43TNWNLDD30 pKa = 3.41DD31 pKa = 5.58DD32 pKa = 5.12PEE34 pKa = 4.47FSAPQEE40 pKa = 4.31SIEE43 pKa = 4.19PSNSIDD49 pKa = 3.37NYY51 pKa = 9.63RR52 pKa = 11.84DD53 pKa = 3.5EE54 pKa = 4.06IVKK57 pKa = 10.36IGVCLSSNNQDD68 pKa = 3.35YY69 pKa = 11.6SFLDD73 pKa = 3.43NQKK76 pKa = 9.89EE77 pKa = 4.06KK78 pKa = 10.96EE79 pKa = 4.04IFLEE83 pKa = 4.67EE84 pKa = 4.0EE85 pKa = 3.92NDD87 pKa = 3.54WEE89 pKa = 4.85VEE91 pKa = 4.29FTKK94 pKa = 10.74KK95 pKa = 10.84LEE97 pKa = 4.07DD98 pKa = 3.46HH99 pKa = 6.44NVPNEE104 pKa = 4.13SEE106 pKa = 4.89LEE108 pKa = 4.06VQINIEE114 pKa = 4.06DD115 pKa = 5.14LIALLHH121 pKa = 5.76FLNVEE126 pKa = 3.71IDD128 pKa = 3.43QYY130 pKa = 11.91NILPLSLQNKK140 pKa = 9.52VILQRR145 pKa = 11.84KK146 pKa = 9.12KK147 pKa = 10.45EE148 pKa = 4.06GKK150 pKa = 9.84SDD152 pKa = 3.49DD153 pKa = 4.06KK154 pKa = 11.25KK155 pKa = 10.39RR156 pKa = 11.84QNGKK160 pKa = 9.83GDD162 pKa = 3.66KK163 pKa = 10.24KK164 pKa = 10.94SCPDD168 pKa = 4.58SKK170 pKa = 11.17NQSNNCQTMNAEE182 pKa = 4.69TNGSTNLGADD192 pKa = 3.65RR193 pKa = 11.84GPNQTEE199 pKa = 4.57DD200 pKa = 3.44NSSMLEE206 pKa = 3.96NDD208 pKa = 3.4SEE210 pKa = 5.74AILDD214 pKa = 3.76SDD216 pKa = 4.4GGEE219 pKa = 3.79LFQFIIKK226 pKa = 9.65TMDD229 pKa = 2.66QGIRR233 pKa = 11.84VKK235 pKa = 10.8KK236 pKa = 9.93KK237 pKa = 9.66FSGKK241 pKa = 8.99KK242 pKa = 9.31VKK244 pKa = 8.69ITRR247 pKa = 11.84EE248 pKa = 4.08NIGLDD253 pKa = 3.08YY254 pKa = 11.02HH255 pKa = 6.95VINGHH260 pKa = 4.91IHH262 pKa = 6.03GKK264 pKa = 7.33SAKK267 pKa = 10.29DD268 pKa = 3.52SIDD271 pKa = 3.28LKK273 pKa = 11.19KK274 pKa = 10.92LMRR277 pKa = 11.84EE278 pKa = 4.37LIKK281 pKa = 10.61KK282 pKa = 7.77GRR284 pKa = 11.84KK285 pKa = 8.52YY286 pKa = 10.86KK287 pKa = 10.57DD288 pKa = 3.21YY289 pKa = 11.42SNQLDD294 pKa = 3.51LSEE297 pKa = 5.02IEE299 pKa = 4.29FF300 pKa = 4.22
Molecular weight: 34.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4HJF7|K4HJF7_ARV Isoform of K4HKI5 Alpha 2 protein OS=Adelaide River virus OX=31612 GN=alpha PE=4 SV=1
MM1 pKa = 7.9ADD3 pKa = 3.31HH4 pKa = 7.08ARR6 pKa = 11.84LISLPSSVQDD16 pKa = 3.38FFNQISSIFKK26 pKa = 10.25SIEE29 pKa = 3.77NFYY32 pKa = 11.17LLYY35 pKa = 9.07KK36 pKa = 9.9TRR38 pKa = 11.84LKK40 pKa = 10.62IFLICTIALIVIVLVAKK57 pKa = 10.32IIKK60 pKa = 8.21YY61 pKa = 7.96TLAFHH66 pKa = 6.95ACIKK70 pKa = 10.08IYY72 pKa = 10.27CKK74 pKa = 10.31PLIKK78 pKa = 8.49TTNCLLKK85 pKa = 10.45LGKK88 pKa = 9.8RR89 pKa = 11.84KK90 pKa = 9.46RR91 pKa = 11.84RR92 pKa = 11.84KK93 pKa = 9.4KK94 pKa = 9.47KK95 pKa = 10.32RR96 pKa = 11.84KK97 pKa = 8.98GKK99 pKa = 9.7SIRR102 pKa = 11.84LKK104 pKa = 10.41EE105 pKa = 3.98LSSS108 pKa = 3.53
MM1 pKa = 7.9ADD3 pKa = 3.31HH4 pKa = 7.08ARR6 pKa = 11.84LISLPSSVQDD16 pKa = 3.38FFNQISSIFKK26 pKa = 10.25SIEE29 pKa = 3.77NFYY32 pKa = 11.17LLYY35 pKa = 9.07KK36 pKa = 9.9TRR38 pKa = 11.84LKK40 pKa = 10.62IFLICTIALIVIVLVAKK57 pKa = 10.32IIKK60 pKa = 8.21YY61 pKa = 7.96TLAFHH66 pKa = 6.95ACIKK70 pKa = 10.08IYY72 pKa = 10.27CKK74 pKa = 10.31PLIKK78 pKa = 8.49TTNCLLKK85 pKa = 10.45LGKK88 pKa = 9.8RR89 pKa = 11.84KK90 pKa = 9.46RR91 pKa = 11.84RR92 pKa = 11.84KK93 pKa = 9.4KK94 pKa = 9.47KK95 pKa = 10.32RR96 pKa = 11.84KK97 pKa = 8.98GKK99 pKa = 9.7SIRR102 pKa = 11.84LKK104 pKa = 10.41EE105 pKa = 3.98LSSS108 pKa = 3.53
Molecular weight: 12.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4719 |
100 |
2144 |
524.3 |
60.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.437 ± 0.265 | 1.759 ± 0.303 |
5.912 ± 0.421 | 6.887 ± 0.655 |
4.153 ± 0.203 | 5.043 ± 0.226 |
2.352 ± 0.252 | 9.176 ± 0.441 |
8.646 ± 0.604 | 9.833 ± 0.651 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.67 ± 0.472 | 7.438 ± 0.482 |
3.285 ± 0.166 | 2.882 ± 0.256 |
4.471 ± 0.269 | 7.819 ± 0.544 |
4.641 ± 0.574 | 4.62 ± 0.45 |
1.441 ± 0.257 | 4.535 ± 0.333 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
