
Sheep faeces associated smacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus sheas1
Average proteome isoelectric point is 5.52
Get precalculated fractions of proteins
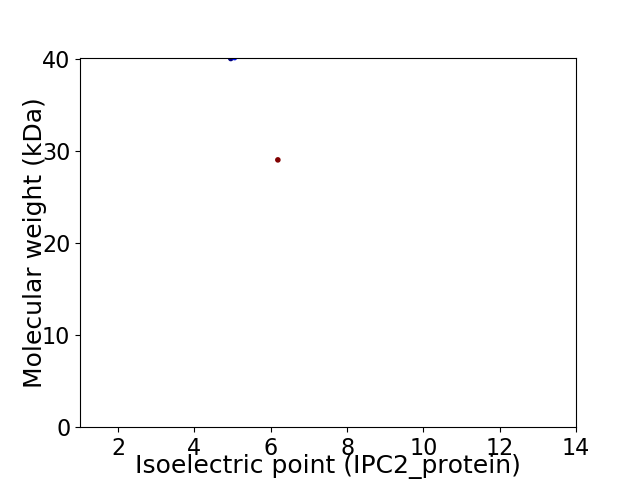
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160HWH4|A0A160HWH4_9VIRU Putative capsid protein OS=Sheep faeces associated smacovirus 1 OX=1843756 PE=4 SV=1
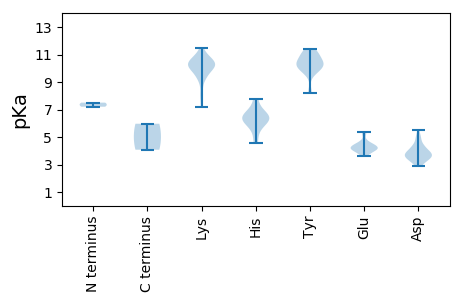
MM1 pKa = 7.2ATNFVKK7 pKa = 10.58CSYY10 pKa = 11.07KK11 pKa = 10.57EE12 pKa = 4.01IIDD15 pKa = 5.19LNTEE19 pKa = 4.29SNKK22 pKa = 8.07VTAIGIHH29 pKa = 6.07TPTGDD34 pKa = 3.69TPRR37 pKa = 11.84KK38 pKa = 8.03MFPGFFKK45 pKa = 10.47QYY47 pKa = 10.24RR48 pKa = 11.84KK49 pKa = 10.15YY50 pKa = 9.94KK51 pKa = 10.19YY52 pKa = 9.94SGASLAFYY60 pKa = 9.82PIAKK64 pKa = 10.0LPADD68 pKa = 4.41PLQVSYY74 pKa = 11.27DD75 pKa = 3.49AGEE78 pKa = 4.23PGIDD82 pKa = 4.05PRR84 pKa = 11.84DD85 pKa = 3.48LANPILVHH93 pKa = 6.23GCHH96 pKa = 6.42GEE98 pKa = 4.37DD99 pKa = 3.86MGQILNKK106 pKa = 10.53LYY108 pKa = 10.93GDD110 pKa = 3.79NNGISDD116 pKa = 4.38SLNLIEE122 pKa = 5.34GGSNIPVVGTDD133 pKa = 2.91PWYY136 pKa = 10.35DD137 pKa = 3.28LMEE140 pKa = 4.52RR141 pKa = 11.84LYY143 pKa = 11.27YY144 pKa = 10.24KK145 pKa = 10.84ALTDD149 pKa = 3.98RR150 pKa = 11.84TWKK153 pKa = 9.48QAGVQRR159 pKa = 11.84GFRR162 pKa = 11.84KK163 pKa = 10.31GGLRR167 pKa = 11.84PLLYY171 pKa = 10.35TLGTNHH177 pKa = 6.7QIMPSSQLSGMVLGDD192 pKa = 3.7DD193 pKa = 4.92GVPKK197 pKa = 10.12MLGGLPTTEE206 pKa = 4.06YY207 pKa = 10.76EE208 pKa = 3.92IDD210 pKa = 3.98EE211 pKa = 4.44EE212 pKa = 5.31DD213 pKa = 5.52EE214 pKa = 4.72PDD216 pKa = 3.81LLNNAVNKK224 pKa = 10.75SNIQFFTPRR233 pKa = 11.84LTRR236 pKa = 11.84LGWMDD241 pKa = 3.35TRR243 pKa = 11.84NVITTNTSQEE253 pKa = 4.48IPDD256 pKa = 3.56SWNADD261 pKa = 3.36TLEE264 pKa = 4.07TLFAQGMVDD273 pKa = 3.37QINWAEE279 pKa = 4.07LPNIYY284 pKa = 9.69MLMILLPPAYY294 pKa = 8.17KK295 pKa = 9.34TEE297 pKa = 3.76MYY299 pKa = 9.86FRR301 pKa = 11.84VVITHH306 pKa = 5.49YY307 pKa = 10.81FKK309 pKa = 11.05FKK311 pKa = 10.0GFRR314 pKa = 11.84GISMMPEE321 pKa = 3.73QTGVPSLVTQEE332 pKa = 4.16WNGSGPEE339 pKa = 4.05TKK341 pKa = 10.16TFDD344 pKa = 4.87DD345 pKa = 4.93LPIDD349 pKa = 4.18PPEE352 pKa = 4.73GGNPRR357 pKa = 11.84AA358 pKa = 4.08
MM1 pKa = 7.2ATNFVKK7 pKa = 10.58CSYY10 pKa = 11.07KK11 pKa = 10.57EE12 pKa = 4.01IIDD15 pKa = 5.19LNTEE19 pKa = 4.29SNKK22 pKa = 8.07VTAIGIHH29 pKa = 6.07TPTGDD34 pKa = 3.69TPRR37 pKa = 11.84KK38 pKa = 8.03MFPGFFKK45 pKa = 10.47QYY47 pKa = 10.24RR48 pKa = 11.84KK49 pKa = 10.15YY50 pKa = 9.94KK51 pKa = 10.19YY52 pKa = 9.94SGASLAFYY60 pKa = 9.82PIAKK64 pKa = 10.0LPADD68 pKa = 4.41PLQVSYY74 pKa = 11.27DD75 pKa = 3.49AGEE78 pKa = 4.23PGIDD82 pKa = 4.05PRR84 pKa = 11.84DD85 pKa = 3.48LANPILVHH93 pKa = 6.23GCHH96 pKa = 6.42GEE98 pKa = 4.37DD99 pKa = 3.86MGQILNKK106 pKa = 10.53LYY108 pKa = 10.93GDD110 pKa = 3.79NNGISDD116 pKa = 4.38SLNLIEE122 pKa = 5.34GGSNIPVVGTDD133 pKa = 2.91PWYY136 pKa = 10.35DD137 pKa = 3.28LMEE140 pKa = 4.52RR141 pKa = 11.84LYY143 pKa = 11.27YY144 pKa = 10.24KK145 pKa = 10.84ALTDD149 pKa = 3.98RR150 pKa = 11.84TWKK153 pKa = 9.48QAGVQRR159 pKa = 11.84GFRR162 pKa = 11.84KK163 pKa = 10.31GGLRR167 pKa = 11.84PLLYY171 pKa = 10.35TLGTNHH177 pKa = 6.7QIMPSSQLSGMVLGDD192 pKa = 3.7DD193 pKa = 4.92GVPKK197 pKa = 10.12MLGGLPTTEE206 pKa = 4.06YY207 pKa = 10.76EE208 pKa = 3.92IDD210 pKa = 3.98EE211 pKa = 4.44EE212 pKa = 5.31DD213 pKa = 5.52EE214 pKa = 4.72PDD216 pKa = 3.81LLNNAVNKK224 pKa = 10.75SNIQFFTPRR233 pKa = 11.84LTRR236 pKa = 11.84LGWMDD241 pKa = 3.35TRR243 pKa = 11.84NVITTNTSQEE253 pKa = 4.48IPDD256 pKa = 3.56SWNADD261 pKa = 3.36TLEE264 pKa = 4.07TLFAQGMVDD273 pKa = 3.37QINWAEE279 pKa = 4.07LPNIYY284 pKa = 9.69MLMILLPPAYY294 pKa = 8.17KK295 pKa = 9.34TEE297 pKa = 3.76MYY299 pKa = 9.86FRR301 pKa = 11.84VVITHH306 pKa = 5.49YY307 pKa = 10.81FKK309 pKa = 11.05FKK311 pKa = 10.0GFRR314 pKa = 11.84GISMMPEE321 pKa = 3.73QTGVPSLVTQEE332 pKa = 4.16WNGSGPEE339 pKa = 4.05TKK341 pKa = 10.16TFDD344 pKa = 4.87DD345 pKa = 4.93LPIDD349 pKa = 4.18PPEE352 pKa = 4.73GGNPRR357 pKa = 11.84AA358 pKa = 4.08
Molecular weight: 40.07 kDa
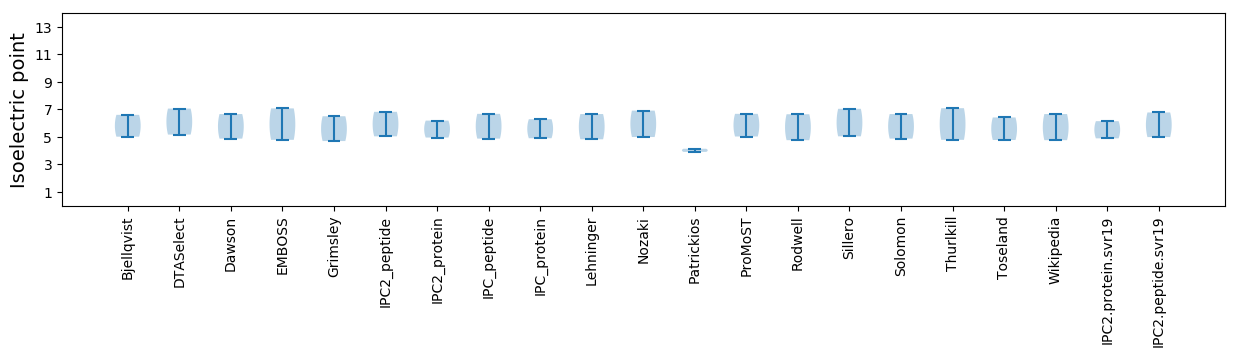
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160HWH4|A0A160HWH4_9VIRU Putative capsid protein OS=Sheep faeces associated smacovirus 1 OX=1843756 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.09KK3 pKa = 10.28YY4 pKa = 10.52VLTVPRR10 pKa = 11.84EE11 pKa = 3.65ADD13 pKa = 3.27LRR15 pKa = 11.84PICRR19 pKa = 11.84MLSDD23 pKa = 3.59AKK25 pKa = 10.41KK26 pKa = 7.15WTIGFEE32 pKa = 4.2VGEE35 pKa = 4.28HH36 pKa = 6.35GFRR39 pKa = 11.84HH40 pKa = 4.55YY41 pKa = 10.59QIRR44 pKa = 11.84LVSSDD49 pKa = 2.98HH50 pKa = 7.77DD51 pKa = 3.66FFEE54 pKa = 4.23WCKK57 pKa = 10.95AFLPTAHH64 pKa = 6.9IEE66 pKa = 4.17EE67 pKa = 4.49ATEE70 pKa = 3.76EE71 pKa = 4.12RR72 pKa = 11.84DD73 pKa = 3.46DD74 pKa = 4.1YY75 pKa = 11.41EE76 pKa = 4.52RR77 pKa = 11.84KK78 pKa = 10.07SGNFLCSDD86 pKa = 3.61DD87 pKa = 3.88TDD89 pKa = 3.6QIRR92 pKa = 11.84QIRR95 pKa = 11.84YY96 pKa = 8.23GDD98 pKa = 3.36LRR100 pKa = 11.84KK101 pKa = 9.42IQKK104 pKa = 10.25KK105 pKa = 9.0ILKK108 pKa = 9.93LADD111 pKa = 3.95DD112 pKa = 3.77QNDD115 pKa = 3.51RR116 pKa = 11.84QISYY120 pKa = 10.26FYY122 pKa = 11.06DD123 pKa = 4.36PDD125 pKa = 4.3GGWAGKK131 pKa = 9.59SWLTIHH137 pKa = 6.29LWEE140 pKa = 5.16RR141 pKa = 11.84GNCFVVPRR149 pKa = 11.84SKK151 pKa = 10.23ATAEE155 pKa = 4.02KK156 pKa = 10.29LSAFICSGYY165 pKa = 10.06KK166 pKa = 9.42AEE168 pKa = 4.35EE169 pKa = 4.43YY170 pKa = 10.35IVIDD174 pKa = 4.53LPRR177 pKa = 11.84APTGDD182 pKa = 2.87IGLYY186 pKa = 10.15EE187 pKa = 4.32LLEE190 pKa = 4.23DD191 pKa = 3.84TKK193 pKa = 11.46NRR195 pKa = 11.84LIFYY199 pKa = 10.34SRR201 pKa = 11.84YY202 pKa = 9.58QPITRR207 pKa = 11.84NIRR210 pKa = 11.84CPNLIVFSNHH220 pKa = 6.4KK221 pKa = 10.4LDD223 pKa = 3.82TKK225 pKa = 10.67RR226 pKa = 11.84LSADD230 pKa = 3.13RR231 pKa = 11.84CQYY234 pKa = 11.09YY235 pKa = 10.51DD236 pKa = 3.76LSVWDD241 pKa = 4.29PLEE244 pKa = 4.01VTLL247 pKa = 5.97
MM1 pKa = 7.5KK2 pKa = 10.09KK3 pKa = 10.28YY4 pKa = 10.52VLTVPRR10 pKa = 11.84EE11 pKa = 3.65ADD13 pKa = 3.27LRR15 pKa = 11.84PICRR19 pKa = 11.84MLSDD23 pKa = 3.59AKK25 pKa = 10.41KK26 pKa = 7.15WTIGFEE32 pKa = 4.2VGEE35 pKa = 4.28HH36 pKa = 6.35GFRR39 pKa = 11.84HH40 pKa = 4.55YY41 pKa = 10.59QIRR44 pKa = 11.84LVSSDD49 pKa = 2.98HH50 pKa = 7.77DD51 pKa = 3.66FFEE54 pKa = 4.23WCKK57 pKa = 10.95AFLPTAHH64 pKa = 6.9IEE66 pKa = 4.17EE67 pKa = 4.49ATEE70 pKa = 3.76EE71 pKa = 4.12RR72 pKa = 11.84DD73 pKa = 3.46DD74 pKa = 4.1YY75 pKa = 11.41EE76 pKa = 4.52RR77 pKa = 11.84KK78 pKa = 10.07SGNFLCSDD86 pKa = 3.61DD87 pKa = 3.88TDD89 pKa = 3.6QIRR92 pKa = 11.84QIRR95 pKa = 11.84YY96 pKa = 8.23GDD98 pKa = 3.36LRR100 pKa = 11.84KK101 pKa = 9.42IQKK104 pKa = 10.25KK105 pKa = 9.0ILKK108 pKa = 9.93LADD111 pKa = 3.95DD112 pKa = 3.77QNDD115 pKa = 3.51RR116 pKa = 11.84QISYY120 pKa = 10.26FYY122 pKa = 11.06DD123 pKa = 4.36PDD125 pKa = 4.3GGWAGKK131 pKa = 9.59SWLTIHH137 pKa = 6.29LWEE140 pKa = 5.16RR141 pKa = 11.84GNCFVVPRR149 pKa = 11.84SKK151 pKa = 10.23ATAEE155 pKa = 4.02KK156 pKa = 10.29LSAFICSGYY165 pKa = 10.06KK166 pKa = 9.42AEE168 pKa = 4.35EE169 pKa = 4.43YY170 pKa = 10.35IVIDD174 pKa = 4.53LPRR177 pKa = 11.84APTGDD182 pKa = 2.87IGLYY186 pKa = 10.15EE187 pKa = 4.32LLEE190 pKa = 4.23DD191 pKa = 3.84TKK193 pKa = 11.46NRR195 pKa = 11.84LIFYY199 pKa = 10.34SRR201 pKa = 11.84YY202 pKa = 9.58QPITRR207 pKa = 11.84NIRR210 pKa = 11.84CPNLIVFSNHH220 pKa = 6.4KK221 pKa = 10.4LDD223 pKa = 3.82TKK225 pKa = 10.67RR226 pKa = 11.84LSADD230 pKa = 3.13RR231 pKa = 11.84CQYY234 pKa = 11.09YY235 pKa = 10.51DD236 pKa = 3.76LSVWDD241 pKa = 4.29PLEE244 pKa = 4.01VTLL247 pKa = 5.97
Molecular weight: 29.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605 |
247 |
358 |
302.5 |
34.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.793 ± 0.267 | 1.488 ± 0.766 |
7.603 ± 0.741 | 5.785 ± 0.394 |
4.132 ± 0.183 | 7.438 ± 1.467 |
1.818 ± 0.347 | 6.612 ± 0.384 |
5.785 ± 0.624 | 9.256 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.645 ± 1.043 | 4.628 ± 1.02 |
6.281 ± 1.269 | 3.471 ± 0.132 |
5.62 ± 1.409 | 5.124 ± 0.309 |
6.446 ± 0.903 | 4.298 ± 0.142 |
1.983 ± 0.253 | 4.793 ± 0.267 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
