
Brachyspira pilosicoli (strain ATCC BAA-1826 / 95/1000)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Brachyspirales; Brachyspiraceae; Brachyspira; Brachyspira pilosicoli
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
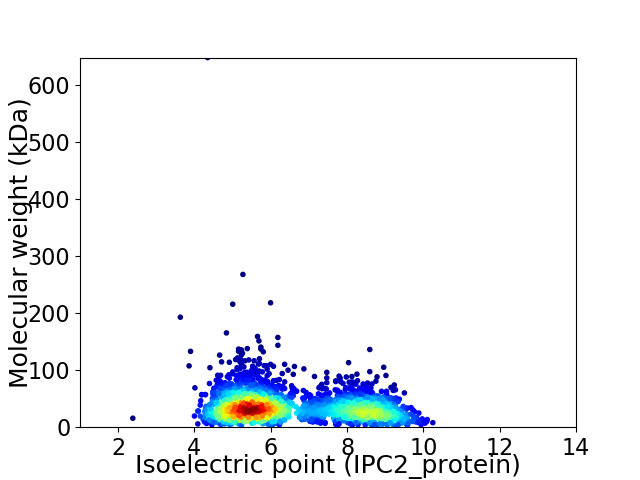
Virtual 2D-PAGE plot for 2301 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8IBM7|D8IBM7_BRAP9 Cell division protein FtsZ OS=Brachyspira pilosicoli (strain ATCC BAA-1826 / 95/1000) OX=759914 GN=ftsZ PE=3 SV=1
MM1 pKa = 7.37FGVFMIKK8 pKa = 10.01KK9 pKa = 9.48VLLLVLFLFVACNNKK24 pKa = 10.28DD25 pKa = 3.76NLDD28 pKa = 3.75KK29 pKa = 11.38NQEE32 pKa = 3.86NTEE35 pKa = 4.2NIEE38 pKa = 3.97NNYY41 pKa = 9.84IDD43 pKa = 4.06TNQNALPEE51 pKa = 4.75TITTTTTTVKK61 pKa = 10.67SDD63 pKa = 2.81GDD65 pKa = 3.84YY66 pKa = 11.58NEE68 pKa = 4.27WFSPYY73 pKa = 10.29TFMKK77 pKa = 10.31NDD79 pKa = 3.81NGSMAQLALNDD90 pKa = 3.91MVLPEE95 pKa = 3.94YY96 pKa = 10.87NIAQFRR102 pKa = 11.84FTYY105 pKa = 9.95IDD107 pKa = 3.35EE108 pKa = 4.44NLIKK112 pKa = 9.97TYY114 pKa = 9.94FYY116 pKa = 11.69SNATIDD122 pKa = 3.65NDD124 pKa = 4.14GADD127 pKa = 4.29MYY129 pKa = 10.97AQSDD133 pKa = 3.26TGYY136 pKa = 8.86TIRR139 pKa = 11.84GNYY142 pKa = 8.02YY143 pKa = 9.55NKK145 pKa = 10.45KK146 pKa = 8.75FTITTKK152 pKa = 9.87IDD154 pKa = 3.44EE155 pKa = 4.42VDD157 pKa = 3.17ISASDD162 pKa = 4.34FISAVSYY169 pKa = 11.06NYY171 pKa = 10.65LYY173 pKa = 10.79TNSSQNDD180 pKa = 3.61SLGNGSTFFYY190 pKa = 10.12KK191 pKa = 10.64DD192 pKa = 3.61SIFVIIPKK200 pKa = 8.77DD201 pKa = 3.66TNNLEE206 pKa = 4.26VYY208 pKa = 10.62DD209 pKa = 5.78KK210 pKa = 10.48ITLDD214 pKa = 3.25INNNIDD220 pKa = 3.64NIVRR224 pKa = 11.84DD225 pKa = 3.54EE226 pKa = 4.16RR227 pKa = 11.84LWLKK231 pKa = 11.12NEE233 pKa = 3.91TLPFFEE239 pKa = 5.1KK240 pKa = 10.79AMSDD244 pKa = 4.88FYY246 pKa = 11.69NDD248 pKa = 3.04WYY250 pKa = 10.14TNSDD254 pKa = 3.3FNYY257 pKa = 10.13EE258 pKa = 4.09YY259 pKa = 10.49IAGKK263 pKa = 10.22DD264 pKa = 2.92ILYY267 pKa = 10.79FNDD270 pKa = 3.25TTISINSYY278 pKa = 9.65SSIYY282 pKa = 9.46MGGAHH287 pKa = 6.44GVYY290 pKa = 10.6NNTSLVYY297 pKa = 10.24SLKK300 pKa = 10.82DD301 pKa = 3.28GEE303 pKa = 4.98KK304 pKa = 10.13ISITNLIKK312 pKa = 10.7DD313 pKa = 4.18FKK315 pKa = 10.89DD316 pKa = 2.99EE317 pKa = 4.05KK318 pKa = 10.96LRR320 pKa = 11.84ISMKK324 pKa = 10.6SKK326 pKa = 10.4LLSISDD332 pKa = 3.75RR333 pKa = 11.84TEE335 pKa = 3.32EE336 pKa = 4.69DD337 pKa = 3.81YY338 pKa = 11.24LVPLDD343 pKa = 5.07EE344 pKa = 4.7ITLEE348 pKa = 4.05DD349 pKa = 3.25TSFYY353 pKa = 10.77VYY355 pKa = 10.66KK356 pKa = 10.85DD357 pKa = 3.59GIHH360 pKa = 5.82FVWPIYY366 pKa = 10.32TITAYY371 pKa = 10.64VQGEE375 pKa = 4.38TEE377 pKa = 3.73IVYY380 pKa = 10.7SFDD383 pKa = 3.63EE384 pKa = 3.87IRR386 pKa = 11.84PFVNEE391 pKa = 3.8EE392 pKa = 3.58YY393 pKa = 10.77LYY395 pKa = 10.5ILEE398 pKa = 4.32EE399 pKa = 4.07
MM1 pKa = 7.37FGVFMIKK8 pKa = 10.01KK9 pKa = 9.48VLLLVLFLFVACNNKK24 pKa = 10.28DD25 pKa = 3.76NLDD28 pKa = 3.75KK29 pKa = 11.38NQEE32 pKa = 3.86NTEE35 pKa = 4.2NIEE38 pKa = 3.97NNYY41 pKa = 9.84IDD43 pKa = 4.06TNQNALPEE51 pKa = 4.75TITTTTTTVKK61 pKa = 10.67SDD63 pKa = 2.81GDD65 pKa = 3.84YY66 pKa = 11.58NEE68 pKa = 4.27WFSPYY73 pKa = 10.29TFMKK77 pKa = 10.31NDD79 pKa = 3.81NGSMAQLALNDD90 pKa = 3.91MVLPEE95 pKa = 3.94YY96 pKa = 10.87NIAQFRR102 pKa = 11.84FTYY105 pKa = 9.95IDD107 pKa = 3.35EE108 pKa = 4.44NLIKK112 pKa = 9.97TYY114 pKa = 9.94FYY116 pKa = 11.69SNATIDD122 pKa = 3.65NDD124 pKa = 4.14GADD127 pKa = 4.29MYY129 pKa = 10.97AQSDD133 pKa = 3.26TGYY136 pKa = 8.86TIRR139 pKa = 11.84GNYY142 pKa = 8.02YY143 pKa = 9.55NKK145 pKa = 10.45KK146 pKa = 8.75FTITTKK152 pKa = 9.87IDD154 pKa = 3.44EE155 pKa = 4.42VDD157 pKa = 3.17ISASDD162 pKa = 4.34FISAVSYY169 pKa = 11.06NYY171 pKa = 10.65LYY173 pKa = 10.79TNSSQNDD180 pKa = 3.61SLGNGSTFFYY190 pKa = 10.12KK191 pKa = 10.64DD192 pKa = 3.61SIFVIIPKK200 pKa = 8.77DD201 pKa = 3.66TNNLEE206 pKa = 4.26VYY208 pKa = 10.62DD209 pKa = 5.78KK210 pKa = 10.48ITLDD214 pKa = 3.25INNNIDD220 pKa = 3.64NIVRR224 pKa = 11.84DD225 pKa = 3.54EE226 pKa = 4.16RR227 pKa = 11.84LWLKK231 pKa = 11.12NEE233 pKa = 3.91TLPFFEE239 pKa = 5.1KK240 pKa = 10.79AMSDD244 pKa = 4.88FYY246 pKa = 11.69NDD248 pKa = 3.04WYY250 pKa = 10.14TNSDD254 pKa = 3.3FNYY257 pKa = 10.13EE258 pKa = 4.09YY259 pKa = 10.49IAGKK263 pKa = 10.22DD264 pKa = 2.92ILYY267 pKa = 10.79FNDD270 pKa = 3.25TTISINSYY278 pKa = 9.65SSIYY282 pKa = 9.46MGGAHH287 pKa = 6.44GVYY290 pKa = 10.6NNTSLVYY297 pKa = 10.24SLKK300 pKa = 10.82DD301 pKa = 3.28GEE303 pKa = 4.98KK304 pKa = 10.13ISITNLIKK312 pKa = 10.7DD313 pKa = 4.18FKK315 pKa = 10.89DD316 pKa = 2.99EE317 pKa = 4.05KK318 pKa = 10.96LRR320 pKa = 11.84ISMKK324 pKa = 10.6SKK326 pKa = 10.4LLSISDD332 pKa = 3.75RR333 pKa = 11.84TEE335 pKa = 3.32EE336 pKa = 4.69DD337 pKa = 3.81YY338 pKa = 11.24LVPLDD343 pKa = 5.07EE344 pKa = 4.7ITLEE348 pKa = 4.05DD349 pKa = 3.25TSFYY353 pKa = 10.77VYY355 pKa = 10.66KK356 pKa = 10.85DD357 pKa = 3.59GIHH360 pKa = 5.82FVWPIYY366 pKa = 10.32TITAYY371 pKa = 10.64VQGEE375 pKa = 4.38TEE377 pKa = 3.73IVYY380 pKa = 10.7SFDD383 pKa = 3.63EE384 pKa = 3.87IRR386 pKa = 11.84PFVNEE391 pKa = 3.8EE392 pKa = 3.58YY393 pKa = 10.77LYY395 pKa = 10.5ILEE398 pKa = 4.32EE399 pKa = 4.07
Molecular weight: 46.42 kDa
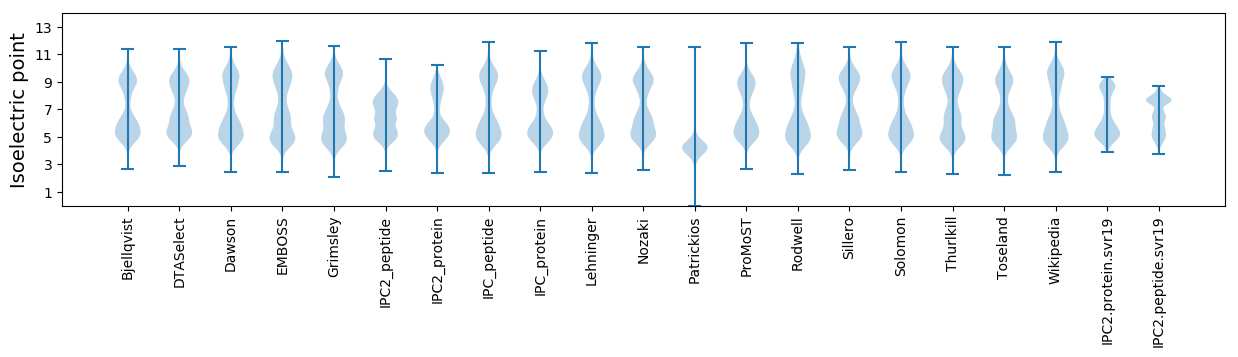
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8ICZ5|D8ICZ5_BRAP9 30S ribosomal protein S10 OS=Brachyspira pilosicoli (strain ATCC BAA-1826 / 95/1000) OX=759914 GN=rpsJ PE=3 SV=1
MM1 pKa = 7.48ARR3 pKa = 11.84LALKK7 pKa = 10.57VKK9 pKa = 8.9ATKK12 pKa = 8.79KK13 pKa = 9.94QKK15 pKa = 10.86YY16 pKa = 6.83KK17 pKa = 8.55TRR19 pKa = 11.84QYY21 pKa = 10.82NRR23 pKa = 11.84CPICGRR29 pKa = 11.84PRR31 pKa = 11.84AYY33 pKa = 9.34IRR35 pKa = 11.84QYY37 pKa = 10.9KK38 pKa = 7.87MCRR41 pKa = 11.84ICFRR45 pKa = 11.84DD46 pKa = 3.63LANKK50 pKa = 10.02GLIPGVTKK58 pKa = 10.8SSWW61 pKa = 2.87
MM1 pKa = 7.48ARR3 pKa = 11.84LALKK7 pKa = 10.57VKK9 pKa = 8.9ATKK12 pKa = 8.79KK13 pKa = 9.94QKK15 pKa = 10.86YY16 pKa = 6.83KK17 pKa = 8.55TRR19 pKa = 11.84QYY21 pKa = 10.82NRR23 pKa = 11.84CPICGRR29 pKa = 11.84PRR31 pKa = 11.84AYY33 pKa = 9.34IRR35 pKa = 11.84QYY37 pKa = 10.9KK38 pKa = 7.87MCRR41 pKa = 11.84ICFRR45 pKa = 11.84DD46 pKa = 3.63LANKK50 pKa = 10.02GLIPGVTKK58 pKa = 10.8SSWW61 pKa = 2.87
Molecular weight: 7.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
762952 |
34 |
5567 |
331.6 |
37.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.917 ± 0.059 | 0.88 ± 0.018 |
5.939 ± 0.057 | 6.857 ± 0.082 |
4.844 ± 0.047 | 5.482 ± 0.071 |
1.293 ± 0.021 | 10.386 ± 0.071 |
8.876 ± 0.053 | 8.765 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 0.025 | 7.591 ± 0.081 |
2.674 ± 0.031 | 2.014 ± 0.023 |
3.155 ± 0.03 | 6.633 ± 0.044 |
4.879 ± 0.041 | 5.768 ± 0.049 |
0.611 ± 0.015 | 4.935 ± 0.053 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
