
Bark beetle-associated genomovirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus denbre4
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
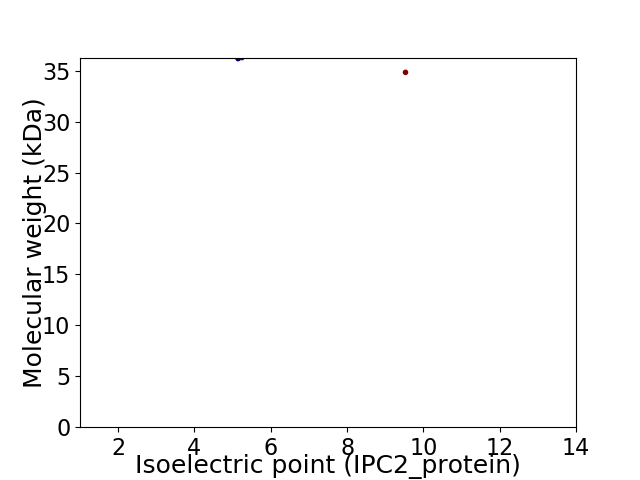
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A344A3Q3|A0A344A3Q3_9VIRU Replication-associated protein OS=Bark beetle-associated genomovirus 4 OX=2230899 PE=3 SV=1
MM1 pKa = 7.94PFHH4 pKa = 7.01CNARR8 pKa = 11.84YY9 pKa = 8.94FLVTYY14 pKa = 9.34SHH16 pKa = 6.96VEE18 pKa = 3.79PLDD21 pKa = 3.65PFALVEE27 pKa = 4.35HH28 pKa = 6.73FGSLGAEE35 pKa = 3.87IIVSLEE41 pKa = 3.82QYY43 pKa = 10.33NATLGVHH50 pKa = 5.02YY51 pKa = 10.39HH52 pKa = 5.64VFADD56 pKa = 4.64FGRR59 pKa = 11.84KK60 pKa = 8.16FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84TDD67 pKa = 2.68IFDD70 pKa = 3.27VDD72 pKa = 3.91GFHH75 pKa = 7.48PNISPSRR82 pKa = 11.84GTPEE86 pKa = 3.98AGYY89 pKa = 10.62DD90 pKa = 3.63YY91 pKa = 10.68AIKK94 pKa = 10.92DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PSGVGATGRR114 pKa = 11.84AAKK117 pKa = 8.68WHH119 pKa = 6.74SILDD123 pKa = 3.54AEE125 pKa = 4.54TRR127 pKa = 11.84DD128 pKa = 3.66EE129 pKa = 5.25FFALCEE135 pKa = 3.97EE136 pKa = 5.06LDD138 pKa = 3.88PEE140 pKa = 4.71RR141 pKa = 11.84LVCSFGQIQKK151 pKa = 9.89FADD154 pKa = 2.98WRR156 pKa = 11.84FAPVAQPYY164 pKa = 9.99ASPDD168 pKa = 3.43GVFDD172 pKa = 4.25LANYY176 pKa = 9.89GDD178 pKa = 3.96LADD181 pKa = 3.68WRR183 pKa = 11.84DD184 pKa = 3.53NFLVADD190 pKa = 3.67TAGRR194 pKa = 11.84SKK196 pKa = 11.1SLILWGPSRR205 pKa = 11.84MGKK208 pKa = 6.7TVWGRR213 pKa = 11.84SLGNHH218 pKa = 7.09LYY220 pKa = 10.71FGGIFSARR228 pKa = 11.84DD229 pKa = 3.28INRR232 pKa = 11.84DD233 pKa = 3.09GVEE236 pKa = 3.89YY237 pKa = 10.85AVFDD241 pKa = 5.39DD242 pKa = 3.46IAGGIKK248 pKa = 10.03FFPRR252 pKa = 11.84FKK254 pKa = 10.79DD255 pKa = 3.12WLGCQMEE262 pKa = 4.36FMVKK266 pKa = 9.32EE267 pKa = 4.5MYY269 pKa = 10.19RR270 pKa = 11.84DD271 pKa = 3.27PHH273 pKa = 6.35LFKK276 pKa = 10.36WGRR279 pKa = 11.84PAIWIANSDD288 pKa = 3.56PRR290 pKa = 11.84HH291 pKa = 6.58DD292 pKa = 3.96MTHH295 pKa = 7.62DD296 pKa = 5.0DD297 pKa = 4.02ITWLEE302 pKa = 4.01ANCIFVEE309 pKa = 4.21ISSPIFHH316 pKa = 7.4ANTEE320 pKa = 4.14
MM1 pKa = 7.94PFHH4 pKa = 7.01CNARR8 pKa = 11.84YY9 pKa = 8.94FLVTYY14 pKa = 9.34SHH16 pKa = 6.96VEE18 pKa = 3.79PLDD21 pKa = 3.65PFALVEE27 pKa = 4.35HH28 pKa = 6.73FGSLGAEE35 pKa = 3.87IIVSLEE41 pKa = 3.82QYY43 pKa = 10.33NATLGVHH50 pKa = 5.02YY51 pKa = 10.39HH52 pKa = 5.64VFADD56 pKa = 4.64FGRR59 pKa = 11.84KK60 pKa = 8.16FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84TDD67 pKa = 2.68IFDD70 pKa = 3.27VDD72 pKa = 3.91GFHH75 pKa = 7.48PNISPSRR82 pKa = 11.84GTPEE86 pKa = 3.98AGYY89 pKa = 10.62DD90 pKa = 3.63YY91 pKa = 10.68AIKK94 pKa = 10.92DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PSGVGATGRR114 pKa = 11.84AAKK117 pKa = 8.68WHH119 pKa = 6.74SILDD123 pKa = 3.54AEE125 pKa = 4.54TRR127 pKa = 11.84DD128 pKa = 3.66EE129 pKa = 5.25FFALCEE135 pKa = 3.97EE136 pKa = 5.06LDD138 pKa = 3.88PEE140 pKa = 4.71RR141 pKa = 11.84LVCSFGQIQKK151 pKa = 9.89FADD154 pKa = 2.98WRR156 pKa = 11.84FAPVAQPYY164 pKa = 9.99ASPDD168 pKa = 3.43GVFDD172 pKa = 4.25LANYY176 pKa = 9.89GDD178 pKa = 3.96LADD181 pKa = 3.68WRR183 pKa = 11.84DD184 pKa = 3.53NFLVADD190 pKa = 3.67TAGRR194 pKa = 11.84SKK196 pKa = 11.1SLILWGPSRR205 pKa = 11.84MGKK208 pKa = 6.7TVWGRR213 pKa = 11.84SLGNHH218 pKa = 7.09LYY220 pKa = 10.71FGGIFSARR228 pKa = 11.84DD229 pKa = 3.28INRR232 pKa = 11.84DD233 pKa = 3.09GVEE236 pKa = 3.89YY237 pKa = 10.85AVFDD241 pKa = 5.39DD242 pKa = 3.46IAGGIKK248 pKa = 10.03FFPRR252 pKa = 11.84FKK254 pKa = 10.79DD255 pKa = 3.12WLGCQMEE262 pKa = 4.36FMVKK266 pKa = 9.32EE267 pKa = 4.5MYY269 pKa = 10.19RR270 pKa = 11.84DD271 pKa = 3.27PHH273 pKa = 6.35LFKK276 pKa = 10.36WGRR279 pKa = 11.84PAIWIANSDD288 pKa = 3.56PRR290 pKa = 11.84HH291 pKa = 6.58DD292 pKa = 3.96MTHH295 pKa = 7.62DD296 pKa = 5.0DD297 pKa = 4.02ITWLEE302 pKa = 4.01ANCIFVEE309 pKa = 4.21ISSPIFHH316 pKa = 7.4ANTEE320 pKa = 4.14
Molecular weight: 36.18 kDa
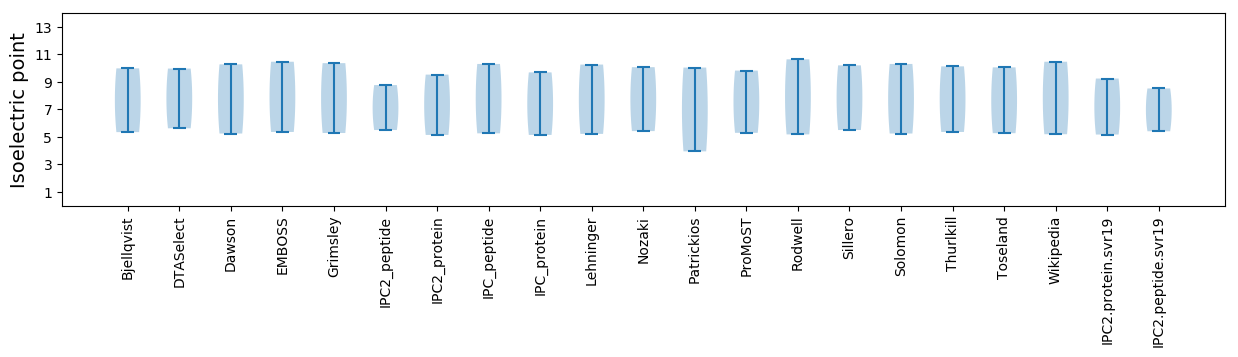
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A344A3Q3|A0A344A3Q3_9VIRU Replication-associated protein OS=Bark beetle-associated genomovirus 4 OX=2230899 PE=3 SV=1
MM1 pKa = 7.39ARR3 pKa = 11.84SRR5 pKa = 11.84YY6 pKa = 9.44YY7 pKa = 10.68GGKK10 pKa = 8.05KK11 pKa = 4.93TTGWANSRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.79SRR24 pKa = 11.84SKK26 pKa = 10.32PRR28 pKa = 11.84RR29 pKa = 11.84SYY31 pKa = 10.73AKK33 pKa = 9.56KK34 pKa = 9.89RR35 pKa = 11.84STFRR39 pKa = 11.84PRR41 pKa = 11.84KK42 pKa = 7.11MSRR45 pKa = 11.84RR46 pKa = 11.84SILNVTSRR54 pKa = 11.84KK55 pKa = 9.78KK56 pKa = 9.85QDD58 pKa = 3.86NMLSWSNKK66 pKa = 8.54NNAGNNQAVATGGLYY81 pKa = 10.82VNGLAGYY88 pKa = 7.97SMCVFCPTARR98 pKa = 11.84SLTTASATNLLVDD111 pKa = 3.53QADD114 pKa = 4.23RR115 pKa = 11.84TSTTCFMRR123 pKa = 11.84GYY125 pKa = 10.29KK126 pKa = 10.04EE127 pKa = 3.71NLRR130 pKa = 11.84IQTSSPLPWLWRR142 pKa = 11.84RR143 pKa = 11.84IVFATKK149 pKa = 10.33GPTFYY154 pKa = 10.96VSSAADD160 pKa = 3.4TPTIKK165 pKa = 9.73FTPYY169 pKa = 10.18SDD171 pKa = 3.5TSVGMARR178 pKa = 11.84LWFDD182 pKa = 3.49LQYY185 pKa = 10.82NSSPATVKK193 pKa = 10.82AMNTIIFKK201 pKa = 10.82GNQDD205 pKa = 3.41QDD207 pKa = 3.47WNDD210 pKa = 3.32VITAKK215 pKa = 10.23VDD217 pKa = 3.47PSRR220 pKa = 11.84ITVMSDD226 pKa = 2.97RR227 pKa = 11.84TCAIKK232 pKa = 8.07TTNSNGHH239 pKa = 4.88FSEE242 pKa = 4.42RR243 pKa = 11.84KK244 pKa = 8.83LWYY247 pKa = 9.61PMNKK251 pKa = 9.49NLVYY255 pKa = 10.91DD256 pKa = 4.4DD257 pKa = 5.44DD258 pKa = 4.18EE259 pKa = 6.9AGAAVASSYY268 pKa = 11.32YY269 pKa = 9.95STDD272 pKa = 3.1AKK274 pKa = 10.71PGMGDD279 pKa = 3.51VYY281 pKa = 11.2VVDD284 pKa = 3.95YY285 pKa = 9.75VVPGVGGATSDD296 pKa = 3.75IINFNATATLYY307 pKa = 9.38WHH309 pKa = 7.04EE310 pKa = 4.29KK311 pKa = 9.3
MM1 pKa = 7.39ARR3 pKa = 11.84SRR5 pKa = 11.84YY6 pKa = 9.44YY7 pKa = 10.68GGKK10 pKa = 8.05KK11 pKa = 4.93TTGWANSRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.79SRR24 pKa = 11.84SKK26 pKa = 10.32PRR28 pKa = 11.84RR29 pKa = 11.84SYY31 pKa = 10.73AKK33 pKa = 9.56KK34 pKa = 9.89RR35 pKa = 11.84STFRR39 pKa = 11.84PRR41 pKa = 11.84KK42 pKa = 7.11MSRR45 pKa = 11.84RR46 pKa = 11.84SILNVTSRR54 pKa = 11.84KK55 pKa = 9.78KK56 pKa = 9.85QDD58 pKa = 3.86NMLSWSNKK66 pKa = 8.54NNAGNNQAVATGGLYY81 pKa = 10.82VNGLAGYY88 pKa = 7.97SMCVFCPTARR98 pKa = 11.84SLTTASATNLLVDD111 pKa = 3.53QADD114 pKa = 4.23RR115 pKa = 11.84TSTTCFMRR123 pKa = 11.84GYY125 pKa = 10.29KK126 pKa = 10.04EE127 pKa = 3.71NLRR130 pKa = 11.84IQTSSPLPWLWRR142 pKa = 11.84RR143 pKa = 11.84IVFATKK149 pKa = 10.33GPTFYY154 pKa = 10.96VSSAADD160 pKa = 3.4TPTIKK165 pKa = 9.73FTPYY169 pKa = 10.18SDD171 pKa = 3.5TSVGMARR178 pKa = 11.84LWFDD182 pKa = 3.49LQYY185 pKa = 10.82NSSPATVKK193 pKa = 10.82AMNTIIFKK201 pKa = 10.82GNQDD205 pKa = 3.41QDD207 pKa = 3.47WNDD210 pKa = 3.32VITAKK215 pKa = 10.23VDD217 pKa = 3.47PSRR220 pKa = 11.84ITVMSDD226 pKa = 2.97RR227 pKa = 11.84TCAIKK232 pKa = 8.07TTNSNGHH239 pKa = 4.88FSEE242 pKa = 4.42RR243 pKa = 11.84KK244 pKa = 8.83LWYY247 pKa = 9.61PMNKK251 pKa = 9.49NLVYY255 pKa = 10.91DD256 pKa = 4.4DD257 pKa = 5.44DD258 pKa = 4.18EE259 pKa = 6.9AGAAVASSYY268 pKa = 11.32YY269 pKa = 9.95STDD272 pKa = 3.1AKK274 pKa = 10.71PGMGDD279 pKa = 3.51VYY281 pKa = 11.2VVDD284 pKa = 3.95YY285 pKa = 9.75VVPGVGGATSDD296 pKa = 3.75IINFNATATLYY307 pKa = 9.38WHH309 pKa = 7.04EE310 pKa = 4.29KK311 pKa = 9.3
Molecular weight: 34.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
631 |
311 |
320 |
315.5 |
35.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.875 ± 0.136 | 1.426 ± 0.099 |
7.29 ± 1.058 | 3.17 ± 1.326 |
5.705 ± 1.753 | 7.448 ± 0.943 |
2.219 ± 1.109 | 4.596 ± 0.745 |
4.913 ± 1.295 | 5.705 ± 0.621 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.536 ± 0.479 | 4.913 ± 1.295 |
4.754 ± 0.404 | 1.902 ± 0.246 |
7.132 ± 0.412 | 7.765 ± 1.55 |
6.498 ± 2.217 | 6.181 ± 0.176 |
2.694 ± 0.086 | 4.279 ± 0.609 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
