
Lachnospiraceae bacterium 3_1_57FAA_CT1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
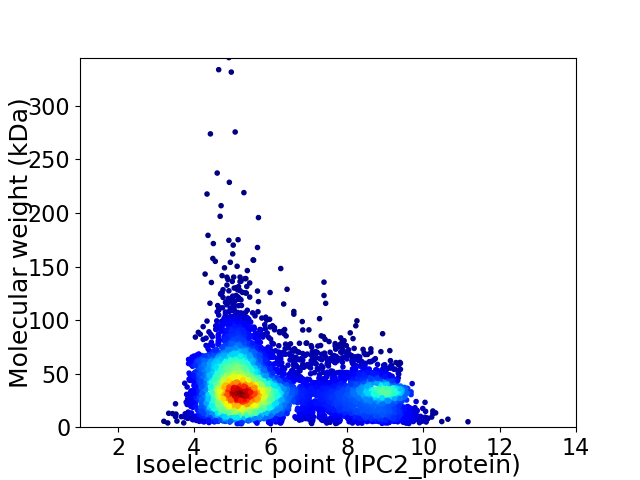
Virtual 2D-PAGE plot for 6505 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F7KHC9|F7KHC9_9FIRM Uncharacterized protein OS=Lachnospiraceae bacterium 3_1_57FAA_CT1 OX=658086 GN=HMPREF0994_05268 PE=4 SV=1
MM1 pKa = 7.22KK2 pKa = 10.16KK3 pKa = 10.17KK4 pKa = 10.68VIAALLTSAMAVSLLAGCGTTVEE27 pKa = 4.69STGSSQSTSTTEE39 pKa = 4.25TKK41 pKa = 8.79EE42 pKa = 3.76TAAPAEE48 pKa = 4.36TEE50 pKa = 4.07TAAVEE55 pKa = 4.34TEE57 pKa = 4.12VAEE60 pKa = 4.52EE61 pKa = 4.45DD62 pKa = 3.88NTDD65 pKa = 3.23LSANITLWTYY75 pKa = 10.65PIGKK79 pKa = 8.33WGDD82 pKa = 3.57SATVDD87 pKa = 4.12SLITSFNAKK96 pKa = 9.57YY97 pKa = 10.28PNINVTVEE105 pKa = 3.79YY106 pKa = 10.84LDD108 pKa = 3.71YY109 pKa = 11.43TNGDD113 pKa = 3.76DD114 pKa = 4.58QVNTAIEE121 pKa = 4.66GGQAPDD127 pKa = 3.74LVMEE131 pKa = 4.7GPEE134 pKa = 3.55RR135 pKa = 11.84LVANWGAKK143 pKa = 9.82GLMVDD148 pKa = 5.07LSDD151 pKa = 5.11IMDD154 pKa = 5.31DD155 pKa = 3.66EE156 pKa = 4.83DD157 pKa = 3.65QSEE160 pKa = 4.74IYY162 pKa = 10.77GSVLAACMTKK172 pKa = 10.21EE173 pKa = 3.98GAVYY177 pKa = 9.21EE178 pKa = 4.46YY179 pKa = 8.93PLCMTAHH186 pKa = 6.68CMAINKK192 pKa = 7.92TVFEE196 pKa = 4.53AAGAMQYY203 pKa = 10.67VDD205 pKa = 4.64EE206 pKa = 6.05DD207 pKa = 3.95SHH209 pKa = 5.84TWTTEE214 pKa = 3.81DD215 pKa = 3.34FFKK218 pKa = 11.22AIDD221 pKa = 3.4AVYY224 pKa = 9.92AHH226 pKa = 6.79TGSTVGAVYY235 pKa = 10.61CSGQGGDD242 pKa = 2.96QGTRR246 pKa = 11.84ALINNLYY253 pKa = 10.69GGTFTDD259 pKa = 4.97DD260 pKa = 3.12LHH262 pKa = 6.33TKK264 pKa = 8.31YY265 pKa = 9.04TADD268 pKa = 3.2SAEE271 pKa = 4.08NVKK274 pKa = 10.48AIQALADD281 pKa = 3.72TDD283 pKa = 3.84GKK285 pKa = 10.69IAFDD289 pKa = 3.65ASIAGGDD296 pKa = 3.84EE297 pKa = 3.95INLFRR302 pKa = 11.84QGVLNVAFCWNIAQQLNSDD321 pKa = 4.14NNDD324 pKa = 3.2AGLTNNGDD332 pKa = 4.11EE333 pKa = 4.43IVFMAFPAEE342 pKa = 4.26KK343 pKa = 10.39ADD345 pKa = 3.43ATQLCGGIWGFGVFDD360 pKa = 4.48NGDD363 pKa = 3.6ADD365 pKa = 5.19RR366 pKa = 11.84IAASKK371 pKa = 10.81LFIKK375 pKa = 10.72YY376 pKa = 8.53MADD379 pKa = 3.59SADD382 pKa = 3.51GTADD386 pKa = 3.39AVLSSTYY393 pKa = 10.12FPVRR397 pKa = 11.84DD398 pKa = 3.77TVEE401 pKa = 4.31GTDD404 pKa = 4.0LTGLYY409 pKa = 10.38NDD411 pKa = 3.66VPTMQEE417 pKa = 3.91YY418 pKa = 10.77SKK420 pKa = 11.23LMQFLGDD427 pKa = 4.01YY428 pKa = 10.0YY429 pKa = 10.98QVTPGWTEE437 pKa = 4.11ARR439 pKa = 11.84TAWWNMLQQVGSGADD454 pKa = 3.16VQTAVDD460 pKa = 4.15EE461 pKa = 4.88FVSTANAAAAAQQ473 pKa = 3.52
MM1 pKa = 7.22KK2 pKa = 10.16KK3 pKa = 10.17KK4 pKa = 10.68VIAALLTSAMAVSLLAGCGTTVEE27 pKa = 4.69STGSSQSTSTTEE39 pKa = 4.25TKK41 pKa = 8.79EE42 pKa = 3.76TAAPAEE48 pKa = 4.36TEE50 pKa = 4.07TAAVEE55 pKa = 4.34TEE57 pKa = 4.12VAEE60 pKa = 4.52EE61 pKa = 4.45DD62 pKa = 3.88NTDD65 pKa = 3.23LSANITLWTYY75 pKa = 10.65PIGKK79 pKa = 8.33WGDD82 pKa = 3.57SATVDD87 pKa = 4.12SLITSFNAKK96 pKa = 9.57YY97 pKa = 10.28PNINVTVEE105 pKa = 3.79YY106 pKa = 10.84LDD108 pKa = 3.71YY109 pKa = 11.43TNGDD113 pKa = 3.76DD114 pKa = 4.58QVNTAIEE121 pKa = 4.66GGQAPDD127 pKa = 3.74LVMEE131 pKa = 4.7GPEE134 pKa = 3.55RR135 pKa = 11.84LVANWGAKK143 pKa = 9.82GLMVDD148 pKa = 5.07LSDD151 pKa = 5.11IMDD154 pKa = 5.31DD155 pKa = 3.66EE156 pKa = 4.83DD157 pKa = 3.65QSEE160 pKa = 4.74IYY162 pKa = 10.77GSVLAACMTKK172 pKa = 10.21EE173 pKa = 3.98GAVYY177 pKa = 9.21EE178 pKa = 4.46YY179 pKa = 8.93PLCMTAHH186 pKa = 6.68CMAINKK192 pKa = 7.92TVFEE196 pKa = 4.53AAGAMQYY203 pKa = 10.67VDD205 pKa = 4.64EE206 pKa = 6.05DD207 pKa = 3.95SHH209 pKa = 5.84TWTTEE214 pKa = 3.81DD215 pKa = 3.34FFKK218 pKa = 11.22AIDD221 pKa = 3.4AVYY224 pKa = 9.92AHH226 pKa = 6.79TGSTVGAVYY235 pKa = 10.61CSGQGGDD242 pKa = 2.96QGTRR246 pKa = 11.84ALINNLYY253 pKa = 10.69GGTFTDD259 pKa = 4.97DD260 pKa = 3.12LHH262 pKa = 6.33TKK264 pKa = 8.31YY265 pKa = 9.04TADD268 pKa = 3.2SAEE271 pKa = 4.08NVKK274 pKa = 10.48AIQALADD281 pKa = 3.72TDD283 pKa = 3.84GKK285 pKa = 10.69IAFDD289 pKa = 3.65ASIAGGDD296 pKa = 3.84EE297 pKa = 3.95INLFRR302 pKa = 11.84QGVLNVAFCWNIAQQLNSDD321 pKa = 4.14NNDD324 pKa = 3.2AGLTNNGDD332 pKa = 4.11EE333 pKa = 4.43IVFMAFPAEE342 pKa = 4.26KK343 pKa = 10.39ADD345 pKa = 3.43ATQLCGGIWGFGVFDD360 pKa = 4.48NGDD363 pKa = 3.6ADD365 pKa = 5.19RR366 pKa = 11.84IAASKK371 pKa = 10.81LFIKK375 pKa = 10.72YY376 pKa = 8.53MADD379 pKa = 3.59SADD382 pKa = 3.51GTADD386 pKa = 3.39AVLSSTYY393 pKa = 10.12FPVRR397 pKa = 11.84DD398 pKa = 3.77TVEE401 pKa = 4.31GTDD404 pKa = 4.0LTGLYY409 pKa = 10.38NDD411 pKa = 3.66VPTMQEE417 pKa = 3.91YY418 pKa = 10.77SKK420 pKa = 11.23LMQFLGDD427 pKa = 4.01YY428 pKa = 10.0YY429 pKa = 10.98QVTPGWTEE437 pKa = 4.11ARR439 pKa = 11.84TAWWNMLQQVGSGADD454 pKa = 3.16VQTAVDD460 pKa = 4.15EE461 pKa = 4.88FVSTANAAAAAQQ473 pKa = 3.52
Molecular weight: 50.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F7KH83|F7KH83_9FIRM ABC transmembrane type-1 domain-containing protein OS=Lachnospiraceae bacterium 3_1_57FAA_CT1 OX=658086 GN=HMPREF0994_05222 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.73MTFQPKK8 pKa = 8.64NRR10 pKa = 11.84QRR12 pKa = 11.84NKK14 pKa = 8.02VHH16 pKa = 6.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.73MTFQPKK8 pKa = 8.64NRR10 pKa = 11.84QRR12 pKa = 11.84NKK14 pKa = 8.02VHH16 pKa = 6.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2265984 |
29 |
3054 |
348.3 |
39.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.42 ± 0.029 | 1.595 ± 0.014 |
5.468 ± 0.024 | 7.745 ± 0.036 |
4.448 ± 0.023 | 7.298 ± 0.025 |
1.686 ± 0.014 | 6.902 ± 0.025 |
6.055 ± 0.024 | 9.472 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.119 ± 0.016 | 4.16 ± 0.019 |
3.614 ± 0.017 | 3.28 ± 0.017 |
4.809 ± 0.025 | 5.903 ± 0.023 |
5.115 ± 0.023 | 6.37 ± 0.023 |
1.188 ± 0.013 | 4.353 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
