
Endomicrobium proavitum
Taxonomy: cellular organisms; Bacteria; Elusimicrobia; Endomicrobia; Endomicrobiales; Endomicrobiaceae; Endomicrobium
Average proteome isoelectric point is 7.49
Get precalculated fractions of proteins
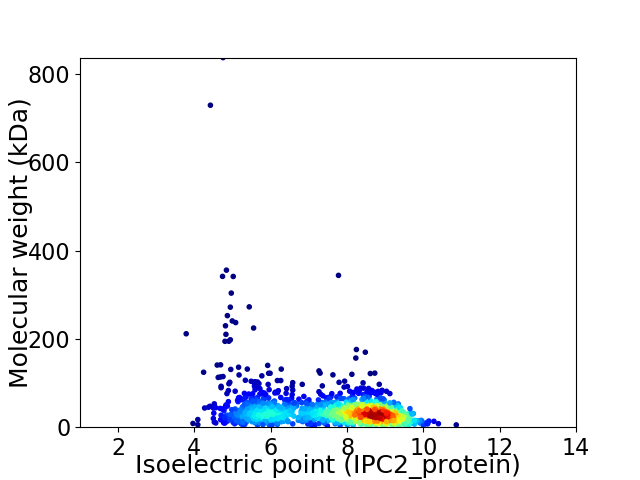
Virtual 2D-PAGE plot for 1336 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G3WJA7|A0A0G3WJA7_9BACT Uncharacterized protein OS=Endomicrobium proavitum OX=1408281 GN=Epro_1044 PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.49KK3 pKa = 10.47SMHH6 pKa = 6.34SKK8 pKa = 9.22WNPFICMLLSFCMTVYY24 pKa = 10.82AFSPVYY30 pKa = 10.18AVNVTQSSDD39 pKa = 3.33FASNWNSAVSPIVVVGAPIDD59 pKa = 3.78SNGNYY64 pKa = 10.5GIILSNYY71 pKa = 8.98LNAIGTNMSLVGYY84 pKa = 9.42SGSPDD89 pKa = 3.25GEE91 pKa = 4.1MLVLGQYY98 pKa = 11.06ALAFAPGWSNDD109 pKa = 2.72INVNISSLSISGATNGAVSFQGNGTGAHH137 pKa = 6.17NLSVSDD143 pKa = 3.15IYY145 pKa = 10.97FYY147 pKa = 11.65NNQFASGNGGAINVGTQIQSVANNKK172 pKa = 9.82FILTDD177 pKa = 3.32NDD179 pKa = 4.15FISNKK184 pKa = 8.8AQNGGAVYY192 pKa = 10.3VSAVNAYY199 pKa = 9.99LSSLLQPVFSVIDD212 pKa = 3.43TTNNNGHH219 pKa = 5.91YY220 pKa = 10.05FAFNQATNGNGGALYY235 pKa = 9.7FASQNAQSTVVNQFTAQGYY254 pKa = 9.61SYY256 pKa = 9.7YY257 pKa = 11.23ANYY260 pKa = 10.89ASGLGGAIYY269 pKa = 10.72NGVSNGNSPAVNEE282 pKa = 4.26FNISSGVFSKK292 pKa = 10.92NQAGSGGAVATTVNGGNTAVNVVINGEE319 pKa = 4.34FYY321 pKa = 11.45SNAATNGGGAVYY333 pKa = 10.66NFVGDD338 pKa = 3.34QSSPYY343 pKa = 9.89GGNINFNVNGIFDD356 pKa = 3.84GNKK359 pKa = 9.45AVNGGAIYY367 pKa = 11.04NNIQPGQATIVTNAGGSFTNNFASGNGGAIYY398 pKa = 8.51NTAGLNTAILNIADD412 pKa = 3.7GTLFWGNGASYY423 pKa = 10.9GGAIFNSGSGVINLNTVSGNAITFGNNYY451 pKa = 9.03AANALNGADD460 pKa = 4.05IYY462 pKa = 11.11QDD464 pKa = 3.19SANAVINITGSGTVNINDD482 pKa = 3.63GFGGIGTINQQTGTTLNLNAPGINSGFTGTFNQAVGSILNASGVMFGGQNNIGGTANINSSQNSFYY548 pKa = 11.08FNANMLSGATLNFTSGSNDD567 pKa = 2.91RR568 pKa = 11.84VSIGVASSVSSPGIRR583 pKa = 11.84FQGTGASVGFNSSVSSGSAYY603 pKa = 11.34NLMQDD608 pKa = 3.79ISNGQSNSVNFNNSNVTFGSTQFTGATAYY637 pKa = 10.28GFYY640 pKa = 10.51NNSVINLADD649 pKa = 3.39SSSNYY654 pKa = 5.97QTYY657 pKa = 10.7NFSTLTTDD665 pKa = 3.55GSGVMNLKK673 pKa = 10.05IGNDD677 pKa = 3.49QNALLSDD684 pKa = 4.01AVNVQNGGGNIGLGKK699 pKa = 10.03IYY701 pKa = 10.47INDD704 pKa = 3.65EE705 pKa = 3.88EE706 pKa = 4.51GLITGYY712 pKa = 9.34MRR714 pKa = 11.84IIYY717 pKa = 8.65GGPLKK722 pKa = 10.3FVNGLTQYY730 pKa = 10.8VATSTGTYY738 pKa = 9.74TITTANDD745 pKa = 3.44YY746 pKa = 10.23YY747 pKa = 11.43VQFNSIPQTPPSGGVVVSTAGAGGDD772 pKa = 3.47TSSTINNIDD781 pKa = 3.36GSTTTVNTNTDD792 pKa = 3.13GSASSTTNNVDD803 pKa = 3.22GSSTTVTTGTDD814 pKa = 3.29GSASVTNTGSGGTDD828 pKa = 3.12TSASTTNPDD837 pKa = 2.89GSTSTTVTNPDD848 pKa = 2.98GSTTTVTTNPDD859 pKa = 2.96GSTSTTNTNAGGTTTSQSTTNPDD882 pKa = 2.79GSTTTTITNPDD893 pKa = 3.51GSTVTTTTNPDD904 pKa = 2.66GSTSVTHH911 pKa = 6.69TDD913 pKa = 2.73STGQSTTVVTNPDD926 pKa = 2.98GSAVITNPDD935 pKa = 3.26SSTTPINPGDD945 pKa = 4.07STSVTTPDD953 pKa = 3.41GSTVDD958 pKa = 3.67VNAGGDD964 pKa = 3.68GTTTTNVTPSGGGNASATTNPDD986 pKa = 2.61GTTTSTTTDD995 pKa = 2.97ADD997 pKa = 3.73GNVINNTTGSDD1008 pKa = 3.72GTSTTTDD1015 pKa = 3.02TTAGGGTAGTTTNPDD1030 pKa = 2.87GTTTSTTTDD1039 pKa = 3.24SNGNTIDD1046 pKa = 4.12SSTSSDD1052 pKa = 3.51GNSTTTNTTPGGTTTITTNPGDD1074 pKa = 3.71NTTVVTDD1081 pKa = 3.43SGGNTTTVITNPDD1094 pKa = 3.39GGGSSASLPDD1104 pKa = 3.39GTTTSTTVDD1113 pKa = 3.57NGDD1116 pKa = 3.2STAQINTPNNQPSLVLVLNDD1136 pKa = 3.19VNAVSVAAFGAGISSSTPRR1155 pKa = 11.84SFQIGANEE1163 pKa = 4.25TYY1165 pKa = 11.2YY1166 pKa = 11.56NDD1168 pKa = 3.92ANLDD1172 pKa = 3.52AMAGGVFTVFGANPGTRR1189 pKa = 11.84TSILSGLNATDD1200 pKa = 3.95LTTQQSLFNIASDD1213 pKa = 4.01SITFTLEE1220 pKa = 3.77DD1221 pKa = 3.53LTITNAYY1228 pKa = 8.13AASGGSVLNMNAANSSAYY1246 pKa = 10.33LGNLLITGNSSGADD1260 pKa = 2.94GGAINVSNGSVYY1272 pKa = 9.21ITGVDD1277 pKa = 4.89FISNSAAGNGGAISNAGGTVVQSGSFIGNSAVNGGAIYY1315 pKa = 8.56NTALAAAPMSIYY1327 pKa = 10.66SATSNLLLSNNTAAALGGAIYY1348 pKa = 10.63NDD1350 pKa = 3.28GFMTIGSATGADD1362 pKa = 3.0IVFSSNTANGVANDD1376 pKa = 4.13IYY1378 pKa = 11.55NSGTLNFASTGGNILITGGIAGSAAGIINKK1408 pKa = 9.74NGLADD1413 pKa = 3.67VVLAATADD1421 pKa = 3.33NSQYY1425 pKa = 11.26LGAFNMTNGIVDD1437 pKa = 3.75ANGKK1441 pKa = 9.51FFGGQSTVQSGILNWNAGASKK1462 pKa = 10.62EE1463 pKa = 4.02EE1464 pKa = 4.17SAILAVTGGVINIYY1478 pKa = 10.99GNLILANANDD1488 pKa = 4.55VIGQYY1493 pKa = 11.63ANLNLFSGGYY1503 pKa = 8.8YY1504 pKa = 9.63EE1505 pKa = 4.28VAGGSVFIDD1514 pKa = 3.6SNDD1517 pKa = 3.43MLVAPDD1523 pKa = 4.2GSYY1526 pKa = 11.58GSAKK1530 pKa = 9.96MSDD1533 pKa = 2.97GHH1535 pKa = 7.17LVLNGQGLVVYY1546 pKa = 10.39SGAYY1550 pKa = 8.12NQTGGVLHH1558 pKa = 6.46VEE1560 pKa = 4.34NQAVTYY1566 pKa = 8.35TIYY1569 pKa = 10.86EE1570 pKa = 4.39GAQSNNGILGGDD1582 pKa = 3.33ISITNARR1589 pKa = 11.84VNVLWYY1595 pKa = 10.47NFNISSGTGSIIPTKK1610 pKa = 10.54GSLSMGDD1617 pKa = 3.02GALFYY1622 pKa = 10.54TVDD1625 pKa = 3.53NTLQTHH1631 pKa = 5.82TFAGDD1636 pKa = 3.42FALFSAVSAFNTAANFEE1653 pKa = 4.27VDD1655 pKa = 5.21LDD1657 pKa = 4.09AQAHH1661 pKa = 6.11KK1662 pKa = 11.0SDD1664 pKa = 3.13ILVFGGTAAPGTIVSQGNPAAGIADD1689 pKa = 3.73VVASSGVVNLSGINFINSPKK1709 pKa = 10.32DD1710 pKa = 3.04AVVPFRR1716 pKa = 11.84IMDD1719 pKa = 4.04APSFDD1724 pKa = 3.47AGIIFSATTKK1734 pKa = 10.28RR1735 pKa = 11.84YY1736 pKa = 6.08YY1737 pKa = 10.31TPIGIYY1743 pKa = 10.16DD1744 pKa = 4.4LYY1746 pKa = 11.62SFGEE1750 pKa = 4.26GNYY1753 pKa = 8.3EE1754 pKa = 4.7LRR1756 pKa = 11.84MDD1758 pKa = 4.02SLNPVTLRR1766 pKa = 11.84AEE1768 pKa = 4.18AAVLAMIDD1776 pKa = 3.57AQNVSNSVLFDD1787 pKa = 3.41HH1788 pKa = 7.08VFFDD1792 pKa = 4.49SNQMLVNANKK1802 pKa = 10.26NEE1804 pKa = 3.68YY1805 pKa = 10.17RR1806 pKa = 11.84FLPRR1810 pKa = 11.84QFLKK1814 pKa = 10.66EE1815 pKa = 4.0NRR1817 pKa = 11.84EE1818 pKa = 3.2KK1819 pKa = 10.95DD1820 pKa = 3.58YY1821 pKa = 10.68WIKK1824 pKa = 9.99TYY1826 pKa = 10.6YY1827 pKa = 8.99EE1828 pKa = 4.15HH1829 pKa = 7.32EE1830 pKa = 4.43AFSVIDD1836 pKa = 3.69NTDD1839 pKa = 3.24LKK1841 pKa = 11.15NNLYY1845 pKa = 10.65GAILGLDD1852 pKa = 4.18FTASDD1857 pKa = 3.59IGEE1860 pKa = 4.19NSYY1863 pKa = 11.21FMPTVFAGYY1872 pKa = 8.33TGASQTLDD1880 pKa = 3.33AASMHH1885 pKa = 5.6QNAVKK1890 pKa = 10.48AGFMASAMVNDD1901 pKa = 4.75VYY1903 pKa = 10.88TVSALVYY1910 pKa = 9.65GGLYY1914 pKa = 8.47QNHH1917 pKa = 6.11MEE1919 pKa = 4.42VDD1921 pKa = 3.69GAEE1924 pKa = 4.35DD1925 pKa = 3.93NLNNWFAGVSVKK1937 pKa = 10.34NAYY1940 pKa = 10.36DD1941 pKa = 3.35MYY1943 pKa = 10.78IDD1945 pKa = 4.23NFVLQPFVTFSYY1957 pKa = 10.79NIYY1960 pKa = 10.59GSQKK1964 pKa = 8.15WNSSYY1969 pKa = 11.41GSIAMQTDD1977 pKa = 3.3NVDD1980 pKa = 3.56GFNASPGVNIIYY1992 pKa = 10.66GLDD1995 pKa = 2.83TWNFVIGEE2003 pKa = 4.24AYY2005 pKa = 10.34VYY2007 pKa = 11.16NLGNKK2012 pKa = 9.51ISGKK2016 pKa = 9.53IGNIEE2021 pKa = 4.91LPDD2024 pKa = 3.94LNNGIGYY2031 pKa = 8.38FEE2033 pKa = 4.22WTAGASKK2040 pKa = 10.7LFSKK2044 pKa = 10.34QLNVWLQAVYY2054 pKa = 8.63KK2055 pKa = 8.42TNRR2058 pKa = 11.84DD2059 pKa = 2.69ADD2061 pKa = 3.6IGVKK2065 pKa = 10.31AGAGWRR2071 pKa = 11.84FF2072 pKa = 3.12
MM1 pKa = 7.42KK2 pKa = 10.49KK3 pKa = 10.47SMHH6 pKa = 6.34SKK8 pKa = 9.22WNPFICMLLSFCMTVYY24 pKa = 10.82AFSPVYY30 pKa = 10.18AVNVTQSSDD39 pKa = 3.33FASNWNSAVSPIVVVGAPIDD59 pKa = 3.78SNGNYY64 pKa = 10.5GIILSNYY71 pKa = 8.98LNAIGTNMSLVGYY84 pKa = 9.42SGSPDD89 pKa = 3.25GEE91 pKa = 4.1MLVLGQYY98 pKa = 11.06ALAFAPGWSNDD109 pKa = 2.72INVNISSLSISGATNGAVSFQGNGTGAHH137 pKa = 6.17NLSVSDD143 pKa = 3.15IYY145 pKa = 10.97FYY147 pKa = 11.65NNQFASGNGGAINVGTQIQSVANNKK172 pKa = 9.82FILTDD177 pKa = 3.32NDD179 pKa = 4.15FISNKK184 pKa = 8.8AQNGGAVYY192 pKa = 10.3VSAVNAYY199 pKa = 9.99LSSLLQPVFSVIDD212 pKa = 3.43TTNNNGHH219 pKa = 5.91YY220 pKa = 10.05FAFNQATNGNGGALYY235 pKa = 9.7FASQNAQSTVVNQFTAQGYY254 pKa = 9.61SYY256 pKa = 9.7YY257 pKa = 11.23ANYY260 pKa = 10.89ASGLGGAIYY269 pKa = 10.72NGVSNGNSPAVNEE282 pKa = 4.26FNISSGVFSKK292 pKa = 10.92NQAGSGGAVATTVNGGNTAVNVVINGEE319 pKa = 4.34FYY321 pKa = 11.45SNAATNGGGAVYY333 pKa = 10.66NFVGDD338 pKa = 3.34QSSPYY343 pKa = 9.89GGNINFNVNGIFDD356 pKa = 3.84GNKK359 pKa = 9.45AVNGGAIYY367 pKa = 11.04NNIQPGQATIVTNAGGSFTNNFASGNGGAIYY398 pKa = 8.51NTAGLNTAILNIADD412 pKa = 3.7GTLFWGNGASYY423 pKa = 10.9GGAIFNSGSGVINLNTVSGNAITFGNNYY451 pKa = 9.03AANALNGADD460 pKa = 4.05IYY462 pKa = 11.11QDD464 pKa = 3.19SANAVINITGSGTVNINDD482 pKa = 3.63GFGGIGTINQQTGTTLNLNAPGINSGFTGTFNQAVGSILNASGVMFGGQNNIGGTANINSSQNSFYY548 pKa = 11.08FNANMLSGATLNFTSGSNDD567 pKa = 2.91RR568 pKa = 11.84VSIGVASSVSSPGIRR583 pKa = 11.84FQGTGASVGFNSSVSSGSAYY603 pKa = 11.34NLMQDD608 pKa = 3.79ISNGQSNSVNFNNSNVTFGSTQFTGATAYY637 pKa = 10.28GFYY640 pKa = 10.51NNSVINLADD649 pKa = 3.39SSSNYY654 pKa = 5.97QTYY657 pKa = 10.7NFSTLTTDD665 pKa = 3.55GSGVMNLKK673 pKa = 10.05IGNDD677 pKa = 3.49QNALLSDD684 pKa = 4.01AVNVQNGGGNIGLGKK699 pKa = 10.03IYY701 pKa = 10.47INDD704 pKa = 3.65EE705 pKa = 3.88EE706 pKa = 4.51GLITGYY712 pKa = 9.34MRR714 pKa = 11.84IIYY717 pKa = 8.65GGPLKK722 pKa = 10.3FVNGLTQYY730 pKa = 10.8VATSTGTYY738 pKa = 9.74TITTANDD745 pKa = 3.44YY746 pKa = 10.23YY747 pKa = 11.43VQFNSIPQTPPSGGVVVSTAGAGGDD772 pKa = 3.47TSSTINNIDD781 pKa = 3.36GSTTTVNTNTDD792 pKa = 3.13GSASSTTNNVDD803 pKa = 3.22GSSTTVTTGTDD814 pKa = 3.29GSASVTNTGSGGTDD828 pKa = 3.12TSASTTNPDD837 pKa = 2.89GSTSTTVTNPDD848 pKa = 2.98GSTTTVTTNPDD859 pKa = 2.96GSTSTTNTNAGGTTTSQSTTNPDD882 pKa = 2.79GSTTTTITNPDD893 pKa = 3.51GSTVTTTTNPDD904 pKa = 2.66GSTSVTHH911 pKa = 6.69TDD913 pKa = 2.73STGQSTTVVTNPDD926 pKa = 2.98GSAVITNPDD935 pKa = 3.26SSTTPINPGDD945 pKa = 4.07STSVTTPDD953 pKa = 3.41GSTVDD958 pKa = 3.67VNAGGDD964 pKa = 3.68GTTTTNVTPSGGGNASATTNPDD986 pKa = 2.61GTTTSTTTDD995 pKa = 2.97ADD997 pKa = 3.73GNVINNTTGSDD1008 pKa = 3.72GTSTTTDD1015 pKa = 3.02TTAGGGTAGTTTNPDD1030 pKa = 2.87GTTTSTTTDD1039 pKa = 3.24SNGNTIDD1046 pKa = 4.12SSTSSDD1052 pKa = 3.51GNSTTTNTTPGGTTTITTNPGDD1074 pKa = 3.71NTTVVTDD1081 pKa = 3.43SGGNTTTVITNPDD1094 pKa = 3.39GGGSSASLPDD1104 pKa = 3.39GTTTSTTVDD1113 pKa = 3.57NGDD1116 pKa = 3.2STAQINTPNNQPSLVLVLNDD1136 pKa = 3.19VNAVSVAAFGAGISSSTPRR1155 pKa = 11.84SFQIGANEE1163 pKa = 4.25TYY1165 pKa = 11.2YY1166 pKa = 11.56NDD1168 pKa = 3.92ANLDD1172 pKa = 3.52AMAGGVFTVFGANPGTRR1189 pKa = 11.84TSILSGLNATDD1200 pKa = 3.95LTTQQSLFNIASDD1213 pKa = 4.01SITFTLEE1220 pKa = 3.77DD1221 pKa = 3.53LTITNAYY1228 pKa = 8.13AASGGSVLNMNAANSSAYY1246 pKa = 10.33LGNLLITGNSSGADD1260 pKa = 2.94GGAINVSNGSVYY1272 pKa = 9.21ITGVDD1277 pKa = 4.89FISNSAAGNGGAISNAGGTVVQSGSFIGNSAVNGGAIYY1315 pKa = 8.56NTALAAAPMSIYY1327 pKa = 10.66SATSNLLLSNNTAAALGGAIYY1348 pKa = 10.63NDD1350 pKa = 3.28GFMTIGSATGADD1362 pKa = 3.0IVFSSNTANGVANDD1376 pKa = 4.13IYY1378 pKa = 11.55NSGTLNFASTGGNILITGGIAGSAAGIINKK1408 pKa = 9.74NGLADD1413 pKa = 3.67VVLAATADD1421 pKa = 3.33NSQYY1425 pKa = 11.26LGAFNMTNGIVDD1437 pKa = 3.75ANGKK1441 pKa = 9.51FFGGQSTVQSGILNWNAGASKK1462 pKa = 10.62EE1463 pKa = 4.02EE1464 pKa = 4.17SAILAVTGGVINIYY1478 pKa = 10.99GNLILANANDD1488 pKa = 4.55VIGQYY1493 pKa = 11.63ANLNLFSGGYY1503 pKa = 8.8YY1504 pKa = 9.63EE1505 pKa = 4.28VAGGSVFIDD1514 pKa = 3.6SNDD1517 pKa = 3.43MLVAPDD1523 pKa = 4.2GSYY1526 pKa = 11.58GSAKK1530 pKa = 9.96MSDD1533 pKa = 2.97GHH1535 pKa = 7.17LVLNGQGLVVYY1546 pKa = 10.39SGAYY1550 pKa = 8.12NQTGGVLHH1558 pKa = 6.46VEE1560 pKa = 4.34NQAVTYY1566 pKa = 8.35TIYY1569 pKa = 10.86EE1570 pKa = 4.39GAQSNNGILGGDD1582 pKa = 3.33ISITNARR1589 pKa = 11.84VNVLWYY1595 pKa = 10.47NFNISSGTGSIIPTKK1610 pKa = 10.54GSLSMGDD1617 pKa = 3.02GALFYY1622 pKa = 10.54TVDD1625 pKa = 3.53NTLQTHH1631 pKa = 5.82TFAGDD1636 pKa = 3.42FALFSAVSAFNTAANFEE1653 pKa = 4.27VDD1655 pKa = 5.21LDD1657 pKa = 4.09AQAHH1661 pKa = 6.11KK1662 pKa = 11.0SDD1664 pKa = 3.13ILVFGGTAAPGTIVSQGNPAAGIADD1689 pKa = 3.73VVASSGVVNLSGINFINSPKK1709 pKa = 10.32DD1710 pKa = 3.04AVVPFRR1716 pKa = 11.84IMDD1719 pKa = 4.04APSFDD1724 pKa = 3.47AGIIFSATTKK1734 pKa = 10.28RR1735 pKa = 11.84YY1736 pKa = 6.08YY1737 pKa = 10.31TPIGIYY1743 pKa = 10.16DD1744 pKa = 4.4LYY1746 pKa = 11.62SFGEE1750 pKa = 4.26GNYY1753 pKa = 8.3EE1754 pKa = 4.7LRR1756 pKa = 11.84MDD1758 pKa = 4.02SLNPVTLRR1766 pKa = 11.84AEE1768 pKa = 4.18AAVLAMIDD1776 pKa = 3.57AQNVSNSVLFDD1787 pKa = 3.41HH1788 pKa = 7.08VFFDD1792 pKa = 4.49SNQMLVNANKK1802 pKa = 10.26NEE1804 pKa = 3.68YY1805 pKa = 10.17RR1806 pKa = 11.84FLPRR1810 pKa = 11.84QFLKK1814 pKa = 10.66EE1815 pKa = 4.0NRR1817 pKa = 11.84EE1818 pKa = 3.2KK1819 pKa = 10.95DD1820 pKa = 3.58YY1821 pKa = 10.68WIKK1824 pKa = 9.99TYY1826 pKa = 10.6YY1827 pKa = 8.99EE1828 pKa = 4.15HH1829 pKa = 7.32EE1830 pKa = 4.43AFSVIDD1836 pKa = 3.69NTDD1839 pKa = 3.24LKK1841 pKa = 11.15NNLYY1845 pKa = 10.65GAILGLDD1852 pKa = 4.18FTASDD1857 pKa = 3.59IGEE1860 pKa = 4.19NSYY1863 pKa = 11.21FMPTVFAGYY1872 pKa = 8.33TGASQTLDD1880 pKa = 3.33AASMHH1885 pKa = 5.6QNAVKK1890 pKa = 10.48AGFMASAMVNDD1901 pKa = 4.75VYY1903 pKa = 10.88TVSALVYY1910 pKa = 9.65GGLYY1914 pKa = 8.47QNHH1917 pKa = 6.11MEE1919 pKa = 4.42VDD1921 pKa = 3.69GAEE1924 pKa = 4.35DD1925 pKa = 3.93NLNNWFAGVSVKK1937 pKa = 10.34NAYY1940 pKa = 10.36DD1941 pKa = 3.35MYY1943 pKa = 10.78IDD1945 pKa = 4.23NFVLQPFVTFSYY1957 pKa = 10.79NIYY1960 pKa = 10.59GSQKK1964 pKa = 8.15WNSSYY1969 pKa = 11.41GSIAMQTDD1977 pKa = 3.3NVDD1980 pKa = 3.56GFNASPGVNIIYY1992 pKa = 10.66GLDD1995 pKa = 2.83TWNFVIGEE2003 pKa = 4.24AYY2005 pKa = 10.34VYY2007 pKa = 11.16NLGNKK2012 pKa = 9.51ISGKK2016 pKa = 9.53IGNIEE2021 pKa = 4.91LPDD2024 pKa = 3.94LNNGIGYY2031 pKa = 8.38FEE2033 pKa = 4.22WTAGASKK2040 pKa = 10.7LFSKK2044 pKa = 10.34QLNVWLQAVYY2054 pKa = 8.63KK2055 pKa = 8.42TNRR2058 pKa = 11.84DD2059 pKa = 2.69ADD2061 pKa = 3.6IGVKK2065 pKa = 10.31AGAGWRR2071 pKa = 11.84FF2072 pKa = 3.12
Molecular weight: 211.73 kDa
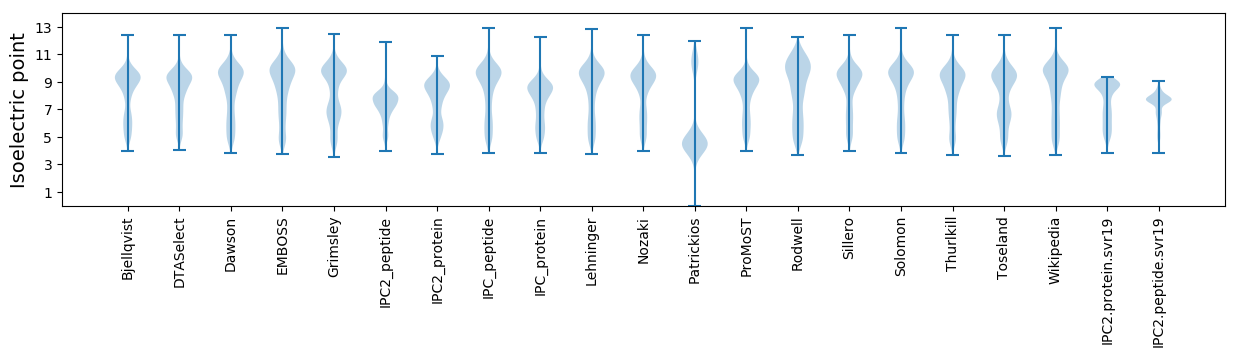
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G3WJQ1|A0A0G3WJQ1_9BACT Uncharacterized protein OS=Endomicrobium proavitum OX=1408281 GN=Epro_1134 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.31QPNKK9 pKa = 9.78DD10 pKa = 3.43RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.9KK14 pKa = 10.69KK15 pKa = 8.91IGFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84RR29 pKa = 11.84VLSARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.2GRR39 pKa = 11.84KK40 pKa = 7.48TISAA44 pKa = 3.98
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.31QPNKK9 pKa = 9.78DD10 pKa = 3.43RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.9KK14 pKa = 10.69KK15 pKa = 8.91IGFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84RR29 pKa = 11.84VLSARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.2GRR39 pKa = 11.84KK40 pKa = 7.48TISAA44 pKa = 3.98
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
479164 |
27 |
8155 |
358.7 |
39.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.874 ± 0.106 | 0.922 ± 0.037 |
4.942 ± 0.064 | 5.789 ± 0.121 |
4.959 ± 0.062 | 7.407 ± 0.183 |
1.31 ± 0.034 | 8.017 ± 0.065 |
8.676 ± 0.187 | 8.321 ± 0.115 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.943 ± 0.039 | 6.093 ± 0.176 |
2.988 ± 0.08 | 2.764 ± 0.044 |
3.191 ± 0.074 | 6.893 ± 0.109 |
5.4 ± 0.15 | 6.879 ± 0.049 |
0.782 ± 0.025 | 3.848 ± 0.055 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
