
Fur seal faeces associated circular DNA virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.6
Get precalculated fractions of proteins
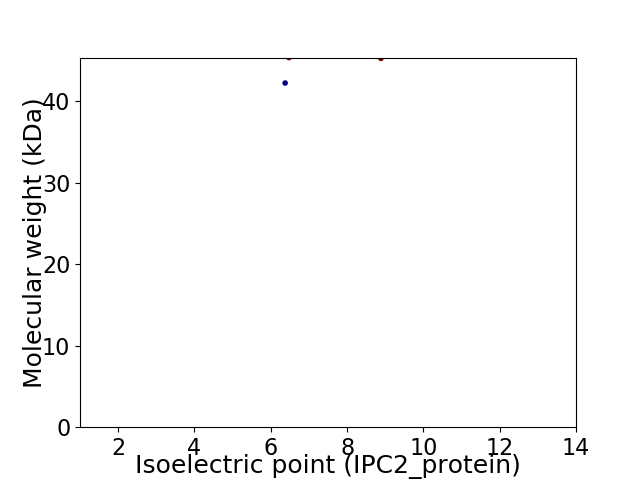
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S4X522|S4X522_9CIRC Capsid protein OS=Fur seal faeces associated circular DNA virus OX=1353241 PE=4 SV=1
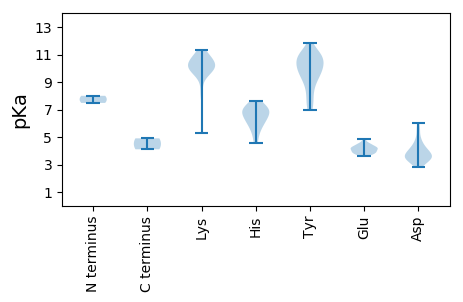
MM1 pKa = 8.0PNPQVKK7 pKa = 8.29MWALTVKK14 pKa = 10.18NWDD17 pKa = 4.06LFDD20 pKa = 4.52HH21 pKa = 6.73PEE23 pKa = 3.87KK24 pKa = 10.98LEE26 pKa = 4.44EE27 pKa = 4.07IGNYY31 pKa = 8.18QDD33 pKa = 4.91EE34 pKa = 4.74DD35 pKa = 3.5GQYY38 pKa = 10.56LVRR41 pKa = 11.84YY42 pKa = 9.4LSIGKK47 pKa = 8.06HH48 pKa = 4.55TGEE51 pKa = 3.85KK52 pKa = 9.0TGYY55 pKa = 7.23QHH57 pKa = 7.03CHH59 pKa = 6.17LNLEE63 pKa = 4.44LEE65 pKa = 4.3KK66 pKa = 10.16RR67 pKa = 11.84QYY69 pKa = 7.13MTWIKK74 pKa = 10.27RR75 pKa = 11.84VLFNIPDD82 pKa = 3.39MHH84 pKa = 6.71CEE86 pKa = 3.68KK87 pKa = 10.63RR88 pKa = 11.84QGTRR92 pKa = 11.84EE93 pKa = 3.83QCDD96 pKa = 3.29TYY98 pKa = 10.93LAKK101 pKa = 10.86DD102 pKa = 3.54GEE104 pKa = 4.49FQVIVNRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84IEE115 pKa = 3.98RR116 pKa = 11.84GHH118 pKa = 6.89RR119 pKa = 11.84SDD121 pKa = 4.94LDD123 pKa = 4.26EE124 pKa = 4.13IHH126 pKa = 7.6DD127 pKa = 4.22MIKK130 pKa = 10.29EE131 pKa = 4.23GKK133 pKa = 9.83DD134 pKa = 3.14LFDD137 pKa = 3.64VYY139 pKa = 10.53EE140 pKa = 4.07SHH142 pKa = 7.11FASTVRR148 pKa = 11.84YY149 pKa = 9.59SSGLEE154 pKa = 3.92KK155 pKa = 11.04YY156 pKa = 9.98IVLHH160 pKa = 6.06DD161 pKa = 3.97SKK163 pKa = 10.76RR164 pKa = 11.84ARR166 pKa = 11.84NSEE169 pKa = 3.89TTAPQVIVYY178 pKa = 8.16VGPAGSGKK186 pKa = 9.7SWHH189 pKa = 6.71CFNDD193 pKa = 3.01EE194 pKa = 4.25DD195 pKa = 4.55YY196 pKa = 11.2KK197 pKa = 10.67EE198 pKa = 4.01SGYY201 pKa = 10.62RR202 pKa = 11.84FPIQMYY208 pKa = 8.87EE209 pKa = 3.61KK210 pKa = 10.72VYY212 pKa = 9.87FDD214 pKa = 3.53GYY216 pKa = 11.03NRR218 pKa = 11.84EE219 pKa = 3.75KK220 pKa = 10.47TIWFDD225 pKa = 3.48EE226 pKa = 4.52FNGRR230 pKa = 11.84SMPFGKK236 pKa = 10.18FCQLADD242 pKa = 4.28RR243 pKa = 11.84FPGIYY248 pKa = 6.94EE249 pKa = 4.19TKK251 pKa = 10.05GGSVLISGLKK261 pKa = 9.94KK262 pKa = 10.24ILISTISYY270 pKa = 7.75PATWWAGSNRR280 pKa = 11.84FNLDD284 pKa = 2.84PDD286 pKa = 3.38QLYY289 pKa = 10.55RR290 pKa = 11.84RR291 pKa = 11.84LTKK294 pKa = 10.12CYY296 pKa = 9.83YY297 pKa = 10.09LGPPRR302 pKa = 11.84RR303 pKa = 11.84SSDD306 pKa = 2.91GSIEE310 pKa = 3.84YY311 pKa = 10.24AIPLEE316 pKa = 4.23FNPKK320 pKa = 9.74EE321 pKa = 4.02LLTKK325 pKa = 9.84EE326 pKa = 3.8QEE328 pKa = 4.54EE329 pKa = 4.04ILKK332 pKa = 10.46RR333 pKa = 11.84HH334 pKa = 5.31VKK336 pKa = 10.35YY337 pKa = 10.55PSDD340 pKa = 4.12YY341 pKa = 11.34NPDD344 pKa = 3.73DD345 pKa = 5.5PILMIRR351 pKa = 11.84DD352 pKa = 3.59RR353 pKa = 11.84LKK355 pKa = 11.12CLLL358 pKa = 4.11
MM1 pKa = 8.0PNPQVKK7 pKa = 8.29MWALTVKK14 pKa = 10.18NWDD17 pKa = 4.06LFDD20 pKa = 4.52HH21 pKa = 6.73PEE23 pKa = 3.87KK24 pKa = 10.98LEE26 pKa = 4.44EE27 pKa = 4.07IGNYY31 pKa = 8.18QDD33 pKa = 4.91EE34 pKa = 4.74DD35 pKa = 3.5GQYY38 pKa = 10.56LVRR41 pKa = 11.84YY42 pKa = 9.4LSIGKK47 pKa = 8.06HH48 pKa = 4.55TGEE51 pKa = 3.85KK52 pKa = 9.0TGYY55 pKa = 7.23QHH57 pKa = 7.03CHH59 pKa = 6.17LNLEE63 pKa = 4.44LEE65 pKa = 4.3KK66 pKa = 10.16RR67 pKa = 11.84QYY69 pKa = 7.13MTWIKK74 pKa = 10.27RR75 pKa = 11.84VLFNIPDD82 pKa = 3.39MHH84 pKa = 6.71CEE86 pKa = 3.68KK87 pKa = 10.63RR88 pKa = 11.84QGTRR92 pKa = 11.84EE93 pKa = 3.83QCDD96 pKa = 3.29TYY98 pKa = 10.93LAKK101 pKa = 10.86DD102 pKa = 3.54GEE104 pKa = 4.49FQVIVNRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84IEE115 pKa = 3.98RR116 pKa = 11.84GHH118 pKa = 6.89RR119 pKa = 11.84SDD121 pKa = 4.94LDD123 pKa = 4.26EE124 pKa = 4.13IHH126 pKa = 7.6DD127 pKa = 4.22MIKK130 pKa = 10.29EE131 pKa = 4.23GKK133 pKa = 9.83DD134 pKa = 3.14LFDD137 pKa = 3.64VYY139 pKa = 10.53EE140 pKa = 4.07SHH142 pKa = 7.11FASTVRR148 pKa = 11.84YY149 pKa = 9.59SSGLEE154 pKa = 3.92KK155 pKa = 11.04YY156 pKa = 9.98IVLHH160 pKa = 6.06DD161 pKa = 3.97SKK163 pKa = 10.76RR164 pKa = 11.84ARR166 pKa = 11.84NSEE169 pKa = 3.89TTAPQVIVYY178 pKa = 8.16VGPAGSGKK186 pKa = 9.7SWHH189 pKa = 6.71CFNDD193 pKa = 3.01EE194 pKa = 4.25DD195 pKa = 4.55YY196 pKa = 11.2KK197 pKa = 10.67EE198 pKa = 4.01SGYY201 pKa = 10.62RR202 pKa = 11.84FPIQMYY208 pKa = 8.87EE209 pKa = 3.61KK210 pKa = 10.72VYY212 pKa = 9.87FDD214 pKa = 3.53GYY216 pKa = 11.03NRR218 pKa = 11.84EE219 pKa = 3.75KK220 pKa = 10.47TIWFDD225 pKa = 3.48EE226 pKa = 4.52FNGRR230 pKa = 11.84SMPFGKK236 pKa = 10.18FCQLADD242 pKa = 4.28RR243 pKa = 11.84FPGIYY248 pKa = 6.94EE249 pKa = 4.19TKK251 pKa = 10.05GGSVLISGLKK261 pKa = 9.94KK262 pKa = 10.24ILISTISYY270 pKa = 7.75PATWWAGSNRR280 pKa = 11.84FNLDD284 pKa = 2.84PDD286 pKa = 3.38QLYY289 pKa = 10.55RR290 pKa = 11.84RR291 pKa = 11.84LTKK294 pKa = 10.12CYY296 pKa = 9.83YY297 pKa = 10.09LGPPRR302 pKa = 11.84RR303 pKa = 11.84SSDD306 pKa = 2.91GSIEE310 pKa = 3.84YY311 pKa = 10.24AIPLEE316 pKa = 4.23FNPKK320 pKa = 9.74EE321 pKa = 4.02LLTKK325 pKa = 9.84EE326 pKa = 3.8QEE328 pKa = 4.54EE329 pKa = 4.04ILKK332 pKa = 10.46RR333 pKa = 11.84HH334 pKa = 5.31VKK336 pKa = 10.35YY337 pKa = 10.55PSDD340 pKa = 4.12YY341 pKa = 11.34NPDD344 pKa = 3.73DD345 pKa = 5.5PILMIRR351 pKa = 11.84DD352 pKa = 3.59RR353 pKa = 11.84LKK355 pKa = 11.12CLLL358 pKa = 4.11
Molecular weight: 42.23 kDa
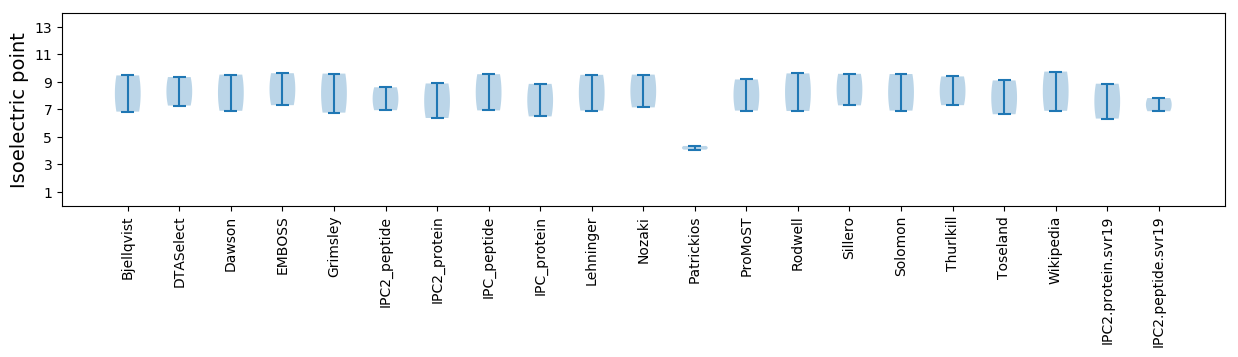
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4X522|S4X522_9CIRC Capsid protein OS=Fur seal faeces associated circular DNA virus OX=1353241 PE=4 SV=1
MM1 pKa = 7.48SPALTLIALVRR12 pKa = 11.84CSLLRR17 pKa = 11.84FFYY20 pKa = 10.94VRR22 pKa = 11.84MVYY25 pKa = 9.84RR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 9.53GKK30 pKa = 9.99RR31 pKa = 11.84YY32 pKa = 7.59SRR34 pKa = 11.84RR35 pKa = 11.84SGRR38 pKa = 11.84SRR40 pKa = 11.84RR41 pKa = 11.84KK42 pKa = 9.38VRR44 pKa = 11.84MSKK47 pKa = 10.27SLYY50 pKa = 8.61KK51 pKa = 9.58RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 9.22RR56 pKa = 11.84RR57 pKa = 11.84YY58 pKa = 8.82RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 9.73SGGLTVSGNRR74 pKa = 11.84EE75 pKa = 3.72SCLEE79 pKa = 3.89YY80 pKa = 11.05SSFIPLATEE89 pKa = 4.21VYY91 pKa = 9.18GSYY94 pKa = 10.42VGSLQDD100 pKa = 5.56GFYY103 pKa = 10.05QWSINPLPAGVCEE116 pKa = 4.1MPGDD120 pKa = 3.63ITYY123 pKa = 9.27TGQWEE128 pKa = 4.2AAGTDD133 pKa = 3.6CFKK136 pKa = 11.02RR137 pKa = 11.84GIGKK141 pKa = 9.59KK142 pKa = 9.9EE143 pKa = 3.74EE144 pKa = 3.97ALMLVDD150 pKa = 5.99LPICISQLSLRR161 pKa = 11.84NLVRR165 pKa = 11.84CDD167 pKa = 3.56TLFEE171 pKa = 4.42LCKK174 pKa = 10.12SYY176 pKa = 10.85RR177 pKa = 11.84IVSVSCTFTVPEE189 pKa = 4.37RR190 pKa = 11.84TSDD193 pKa = 3.89GPNHH197 pKa = 6.34NLYY200 pKa = 11.12LMWTHH205 pKa = 7.19LPKK208 pKa = 10.61CRR210 pKa = 11.84AADD213 pKa = 3.64AEE215 pKa = 4.87SCFGMVCKK223 pKa = 10.2EE224 pKa = 3.88VVNIEE229 pKa = 4.24GQYY232 pKa = 10.22SPSVSGWNWIANPADD247 pKa = 3.22IAEE250 pKa = 4.54ACSCDD255 pKa = 3.69GIEE258 pKa = 4.24NGRR261 pKa = 11.84NGWQRR266 pKa = 11.84KK267 pKa = 5.3QLAYY271 pKa = 10.37NSPVTISWRR280 pKa = 11.84PRR282 pKa = 11.84HH283 pKa = 5.58AKK285 pKa = 10.04IIASHH290 pKa = 6.11EE291 pKa = 4.09NYY293 pKa = 9.52IDD295 pKa = 3.33SGYY298 pKa = 10.98AHH300 pKa = 7.14NAAQTYY306 pKa = 10.72NKK308 pKa = 9.58VLPNFASDD316 pKa = 3.53LKK318 pKa = 10.36MSRR321 pKa = 11.84GYY323 pKa = 10.98LPTDD327 pKa = 3.07IDD329 pKa = 3.45DD330 pKa = 3.93RR331 pKa = 11.84ARR333 pKa = 11.84TEE335 pKa = 3.67RR336 pKa = 11.84QYY338 pKa = 11.82WMGPCIRR345 pKa = 11.84LIDD348 pKa = 4.0GDD350 pKa = 3.98IKK352 pKa = 11.05ASEE355 pKa = 4.24PVPEE359 pKa = 4.31TSALNIFDD367 pKa = 4.0RR368 pKa = 11.84YY369 pKa = 9.79GIRR372 pKa = 11.84VTTTIKK378 pKa = 10.6VRR380 pKa = 11.84FKK382 pKa = 11.34GMNTSDD388 pKa = 4.61PIFPEE393 pKa = 4.2YY394 pKa = 9.94QAA396 pKa = 4.91
MM1 pKa = 7.48SPALTLIALVRR12 pKa = 11.84CSLLRR17 pKa = 11.84FFYY20 pKa = 10.94VRR22 pKa = 11.84MVYY25 pKa = 9.84RR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 9.53GKK30 pKa = 9.99RR31 pKa = 11.84YY32 pKa = 7.59SRR34 pKa = 11.84RR35 pKa = 11.84SGRR38 pKa = 11.84SRR40 pKa = 11.84RR41 pKa = 11.84KK42 pKa = 9.38VRR44 pKa = 11.84MSKK47 pKa = 10.27SLYY50 pKa = 8.61KK51 pKa = 9.58RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 9.22RR56 pKa = 11.84RR57 pKa = 11.84YY58 pKa = 8.82RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 9.73SGGLTVSGNRR74 pKa = 11.84EE75 pKa = 3.72SCLEE79 pKa = 3.89YY80 pKa = 11.05SSFIPLATEE89 pKa = 4.21VYY91 pKa = 9.18GSYY94 pKa = 10.42VGSLQDD100 pKa = 5.56GFYY103 pKa = 10.05QWSINPLPAGVCEE116 pKa = 4.1MPGDD120 pKa = 3.63ITYY123 pKa = 9.27TGQWEE128 pKa = 4.2AAGTDD133 pKa = 3.6CFKK136 pKa = 11.02RR137 pKa = 11.84GIGKK141 pKa = 9.59KK142 pKa = 9.9EE143 pKa = 3.74EE144 pKa = 3.97ALMLVDD150 pKa = 5.99LPICISQLSLRR161 pKa = 11.84NLVRR165 pKa = 11.84CDD167 pKa = 3.56TLFEE171 pKa = 4.42LCKK174 pKa = 10.12SYY176 pKa = 10.85RR177 pKa = 11.84IVSVSCTFTVPEE189 pKa = 4.37RR190 pKa = 11.84TSDD193 pKa = 3.89GPNHH197 pKa = 6.34NLYY200 pKa = 11.12LMWTHH205 pKa = 7.19LPKK208 pKa = 10.61CRR210 pKa = 11.84AADD213 pKa = 3.64AEE215 pKa = 4.87SCFGMVCKK223 pKa = 10.2EE224 pKa = 3.88VVNIEE229 pKa = 4.24GQYY232 pKa = 10.22SPSVSGWNWIANPADD247 pKa = 3.22IAEE250 pKa = 4.54ACSCDD255 pKa = 3.69GIEE258 pKa = 4.24NGRR261 pKa = 11.84NGWQRR266 pKa = 11.84KK267 pKa = 5.3QLAYY271 pKa = 10.37NSPVTISWRR280 pKa = 11.84PRR282 pKa = 11.84HH283 pKa = 5.58AKK285 pKa = 10.04IIASHH290 pKa = 6.11EE291 pKa = 4.09NYY293 pKa = 9.52IDD295 pKa = 3.33SGYY298 pKa = 10.98AHH300 pKa = 7.14NAAQTYY306 pKa = 10.72NKK308 pKa = 9.58VLPNFASDD316 pKa = 3.53LKK318 pKa = 10.36MSRR321 pKa = 11.84GYY323 pKa = 10.98LPTDD327 pKa = 3.07IDD329 pKa = 3.45DD330 pKa = 3.93RR331 pKa = 11.84ARR333 pKa = 11.84TEE335 pKa = 3.67RR336 pKa = 11.84QYY338 pKa = 11.82WMGPCIRR345 pKa = 11.84LIDD348 pKa = 4.0GDD350 pKa = 3.98IKK352 pKa = 11.05ASEE355 pKa = 4.24PVPEE359 pKa = 4.31TSALNIFDD367 pKa = 4.0RR368 pKa = 11.84YY369 pKa = 9.79GIRR372 pKa = 11.84VTTTIKK378 pKa = 10.6VRR380 pKa = 11.84FKK382 pKa = 11.34GMNTSDD388 pKa = 4.61PIFPEE393 pKa = 4.2YY394 pKa = 9.94QAA396 pKa = 4.91
Molecular weight: 45.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
754 |
358 |
396 |
377.0 |
43.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.642 ± 1.204 | 2.785 ± 0.541 |
5.703 ± 0.834 | 6.233 ± 1.034 |
3.581 ± 0.397 | 6.764 ± 0.039 |
2.122 ± 0.619 | 6.101 ± 0.029 |
5.968 ± 1.025 | 7.692 ± 0.448 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.387 ± 0.099 | 4.111 ± 0.131 |
5.04 ± 0.008 | 3.05 ± 0.378 |
8.621 ± 1.067 | 7.56 ± 1.103 |
4.642 ± 0.294 | 4.907 ± 0.467 |
1.989 ± 0.022 | 6.101 ± 0.211 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
