
Banna virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Seadornavirus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
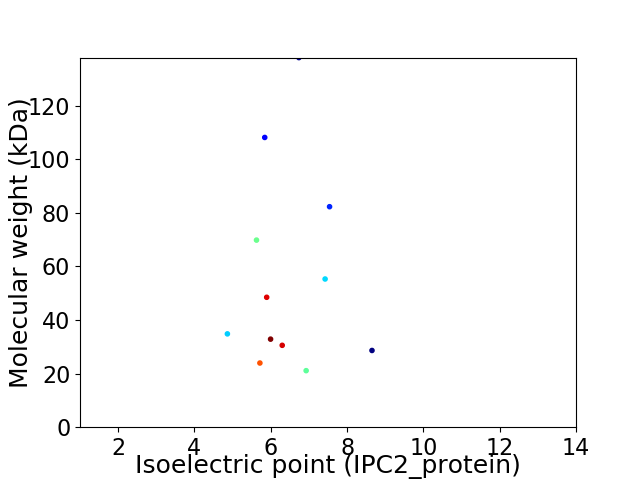
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B4Y052|B4Y052_9REOV VP8 OS=Banna virus OX=77763 PE=4 SV=1
MM1 pKa = 7.58NNGQATITRR10 pKa = 11.84IGRR13 pKa = 11.84NFEE16 pKa = 3.56IRR18 pKa = 11.84CRR20 pKa = 11.84HH21 pKa = 6.31LDD23 pKa = 3.38RR24 pKa = 11.84DD25 pKa = 3.97YY26 pKa = 11.43TLPLPNATSNDD37 pKa = 3.34NFLDD41 pKa = 4.43CIKK44 pKa = 10.76FITEE48 pKa = 4.04CVGFNYY54 pKa = 9.95VSSGFKK60 pKa = 10.61LSIDD64 pKa = 3.39INDD67 pKa = 3.76FQHH70 pKa = 6.83FNGNSTLLIGKK81 pKa = 6.81TKK83 pKa = 10.22IGPLILKK90 pKa = 9.61KK91 pKa = 10.56VRR93 pKa = 11.84SLPCCNDD100 pKa = 2.73ALFRR104 pKa = 11.84NEE106 pKa = 3.65FRR108 pKa = 11.84ILAKK112 pKa = 9.4MHH114 pKa = 6.73GILRR118 pKa = 11.84LKK120 pKa = 10.8NDD122 pKa = 3.56INGHH126 pKa = 5.51KK127 pKa = 10.31YY128 pKa = 9.91GVILEE133 pKa = 4.27RR134 pKa = 11.84CYY136 pKa = 10.84KK137 pKa = 10.5PNINFCNFVTAINDD151 pKa = 3.58LEE153 pKa = 4.48VFHH156 pKa = 7.05SSNPHH161 pKa = 5.99FLHH164 pKa = 7.5GDD166 pKa = 3.65ANPDD170 pKa = 3.6NIMSDD175 pKa = 2.81SDD177 pKa = 4.39GYY179 pKa = 11.32LKK181 pKa = 10.89LVDD184 pKa = 4.19PVCLLEE190 pKa = 4.24NQVNMVNVDD199 pKa = 3.77YY200 pKa = 11.23EE201 pKa = 4.54SLTQEE206 pKa = 3.9AEE208 pKa = 3.77KK209 pKa = 10.82KK210 pKa = 10.5VFVKK214 pKa = 10.73SLLLLVEE221 pKa = 4.43KK222 pKa = 10.44QLSASADD229 pKa = 3.63EE230 pKa = 4.89IYY232 pKa = 11.55VNLKK236 pKa = 7.26EE237 pKa = 4.53TNPSFNLEE245 pKa = 4.29CGLKK249 pKa = 9.49LTDD252 pKa = 4.93LLDD255 pKa = 5.21SIDD258 pKa = 3.76THH260 pKa = 8.08NSIHH264 pKa = 6.23WKK266 pKa = 10.7SMLNYY271 pKa = 9.51QPIMPEE277 pKa = 3.89LSVLNDD283 pKa = 3.14LTYY286 pKa = 11.22YY287 pKa = 9.89DD288 pKa = 3.87TGDD291 pKa = 3.55VKK293 pKa = 11.36DD294 pKa = 5.33LVTEE298 pKa = 4.51DD299 pKa = 5.48LDD301 pKa = 6.27DD302 pKa = 4.69EE303 pKa = 5.88DD304 pKa = 5.88DD305 pKa = 3.7VV306 pKa = 4.58
MM1 pKa = 7.58NNGQATITRR10 pKa = 11.84IGRR13 pKa = 11.84NFEE16 pKa = 3.56IRR18 pKa = 11.84CRR20 pKa = 11.84HH21 pKa = 6.31LDD23 pKa = 3.38RR24 pKa = 11.84DD25 pKa = 3.97YY26 pKa = 11.43TLPLPNATSNDD37 pKa = 3.34NFLDD41 pKa = 4.43CIKK44 pKa = 10.76FITEE48 pKa = 4.04CVGFNYY54 pKa = 9.95VSSGFKK60 pKa = 10.61LSIDD64 pKa = 3.39INDD67 pKa = 3.76FQHH70 pKa = 6.83FNGNSTLLIGKK81 pKa = 6.81TKK83 pKa = 10.22IGPLILKK90 pKa = 9.61KK91 pKa = 10.56VRR93 pKa = 11.84SLPCCNDD100 pKa = 2.73ALFRR104 pKa = 11.84NEE106 pKa = 3.65FRR108 pKa = 11.84ILAKK112 pKa = 9.4MHH114 pKa = 6.73GILRR118 pKa = 11.84LKK120 pKa = 10.8NDD122 pKa = 3.56INGHH126 pKa = 5.51KK127 pKa = 10.31YY128 pKa = 9.91GVILEE133 pKa = 4.27RR134 pKa = 11.84CYY136 pKa = 10.84KK137 pKa = 10.5PNINFCNFVTAINDD151 pKa = 3.58LEE153 pKa = 4.48VFHH156 pKa = 7.05SSNPHH161 pKa = 5.99FLHH164 pKa = 7.5GDD166 pKa = 3.65ANPDD170 pKa = 3.6NIMSDD175 pKa = 2.81SDD177 pKa = 4.39GYY179 pKa = 11.32LKK181 pKa = 10.89LVDD184 pKa = 4.19PVCLLEE190 pKa = 4.24NQVNMVNVDD199 pKa = 3.77YY200 pKa = 11.23EE201 pKa = 4.54SLTQEE206 pKa = 3.9AEE208 pKa = 3.77KK209 pKa = 10.82KK210 pKa = 10.5VFVKK214 pKa = 10.73SLLLLVEE221 pKa = 4.43KK222 pKa = 10.44QLSASADD229 pKa = 3.63EE230 pKa = 4.89IYY232 pKa = 11.55VNLKK236 pKa = 7.26EE237 pKa = 4.53TNPSFNLEE245 pKa = 4.29CGLKK249 pKa = 9.49LTDD252 pKa = 4.93LLDD255 pKa = 5.21SIDD258 pKa = 3.76THH260 pKa = 8.08NSIHH264 pKa = 6.23WKK266 pKa = 10.7SMLNYY271 pKa = 9.51QPIMPEE277 pKa = 3.89LSVLNDD283 pKa = 3.14LTYY286 pKa = 11.22YY287 pKa = 9.89DD288 pKa = 3.87TGDD291 pKa = 3.55VKK293 pKa = 11.36DD294 pKa = 5.33LVTEE298 pKa = 4.51DD299 pKa = 5.48LDD301 pKa = 6.27DD302 pKa = 4.69EE303 pKa = 5.88DD304 pKa = 5.88DD305 pKa = 3.7VV306 pKa = 4.58
Molecular weight: 34.82 kDa
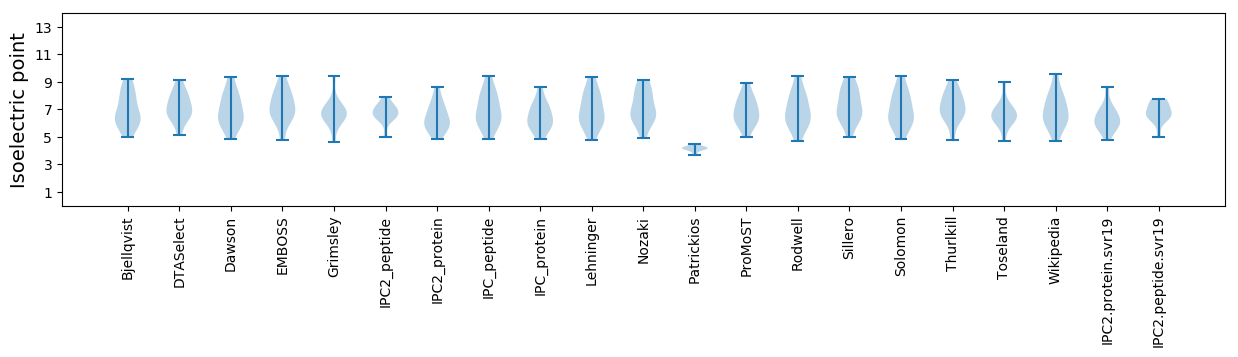
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B4Y055|B4Y055_9REOV VP11 OS=Banna virus OX=77763 PE=4 SV=1
MM1 pKa = 7.93DD2 pKa = 4.23VLSKK6 pKa = 11.18SSLKK10 pKa = 10.48EE11 pKa = 3.7LLAHH15 pKa = 7.03LEE17 pKa = 4.06KK18 pKa = 10.52TPLEE22 pKa = 3.95EE23 pKa = 5.08AISYY27 pKa = 10.23KK28 pKa = 10.25IGTIPYY34 pKa = 8.23QNVFISRR41 pKa = 11.84NEE43 pKa = 4.28HH44 pKa = 5.74YY45 pKa = 10.61NQLYY49 pKa = 10.0PDD51 pKa = 3.95TTSLIDD57 pKa = 3.54GVSRR61 pKa = 11.84EE62 pKa = 4.0GQRR65 pKa = 11.84NVNGLIMSIISYY77 pKa = 9.13VVSGSGHH84 pKa = 6.27YY85 pKa = 9.77IPNIGSMLLRR95 pKa = 11.84RR96 pKa = 11.84SILDD100 pKa = 3.1ILTKK104 pKa = 10.3HH105 pKa = 6.06DD106 pKa = 3.68TGLVTNNLNYY116 pKa = 10.81GIIARR121 pKa = 11.84NLTVNKK127 pKa = 9.03MNCEE131 pKa = 3.59QRR133 pKa = 11.84KK134 pKa = 9.66RR135 pKa = 11.84MLICFKK141 pKa = 10.89LLAYY145 pKa = 9.8KK146 pKa = 10.52DD147 pKa = 4.31GNQNDD152 pKa = 3.94YY153 pKa = 11.14EE154 pKa = 4.51VYY156 pKa = 10.56LNQNIPLKK164 pKa = 10.6QIAPSFIPNDD174 pKa = 3.12MRR176 pKa = 11.84TVINNNDD183 pKa = 3.09QLGIVGIPVYY193 pKa = 10.52RR194 pKa = 11.84LTQSTEE200 pKa = 3.33LSIRR204 pKa = 11.84DD205 pKa = 4.2DD206 pKa = 3.48NAKK209 pKa = 10.12SYY211 pKa = 11.21KK212 pKa = 10.04LGYY215 pKa = 9.08VDD217 pKa = 4.72WYY219 pKa = 10.85NSNAFLRR226 pKa = 11.84EE227 pKa = 3.87RR228 pKa = 11.84NDD230 pKa = 3.35FNLLKK235 pKa = 10.89LKK237 pKa = 10.45DD238 pKa = 3.42RR239 pKa = 11.84NTRR242 pKa = 11.84YY243 pKa = 10.27GRR245 pKa = 11.84LIGWW249 pKa = 3.97
MM1 pKa = 7.93DD2 pKa = 4.23VLSKK6 pKa = 11.18SSLKK10 pKa = 10.48EE11 pKa = 3.7LLAHH15 pKa = 7.03LEE17 pKa = 4.06KK18 pKa = 10.52TPLEE22 pKa = 3.95EE23 pKa = 5.08AISYY27 pKa = 10.23KK28 pKa = 10.25IGTIPYY34 pKa = 8.23QNVFISRR41 pKa = 11.84NEE43 pKa = 4.28HH44 pKa = 5.74YY45 pKa = 10.61NQLYY49 pKa = 10.0PDD51 pKa = 3.95TTSLIDD57 pKa = 3.54GVSRR61 pKa = 11.84EE62 pKa = 4.0GQRR65 pKa = 11.84NVNGLIMSIISYY77 pKa = 9.13VVSGSGHH84 pKa = 6.27YY85 pKa = 9.77IPNIGSMLLRR95 pKa = 11.84RR96 pKa = 11.84SILDD100 pKa = 3.1ILTKK104 pKa = 10.3HH105 pKa = 6.06DD106 pKa = 3.68TGLVTNNLNYY116 pKa = 10.81GIIARR121 pKa = 11.84NLTVNKK127 pKa = 9.03MNCEE131 pKa = 3.59QRR133 pKa = 11.84KK134 pKa = 9.66RR135 pKa = 11.84MLICFKK141 pKa = 10.89LLAYY145 pKa = 9.8KK146 pKa = 10.52DD147 pKa = 4.31GNQNDD152 pKa = 3.94YY153 pKa = 11.14EE154 pKa = 4.51VYY156 pKa = 10.56LNQNIPLKK164 pKa = 10.6QIAPSFIPNDD174 pKa = 3.12MRR176 pKa = 11.84TVINNNDD183 pKa = 3.09QLGIVGIPVYY193 pKa = 10.52RR194 pKa = 11.84LTQSTEE200 pKa = 3.33LSIRR204 pKa = 11.84DD205 pKa = 4.2DD206 pKa = 3.48NAKK209 pKa = 10.12SYY211 pKa = 11.21KK212 pKa = 10.04LGYY215 pKa = 9.08VDD217 pKa = 4.72WYY219 pKa = 10.85NSNAFLRR226 pKa = 11.84EE227 pKa = 3.87RR228 pKa = 11.84NDD230 pKa = 3.35FNLLKK235 pKa = 10.89LKK237 pKa = 10.45DD238 pKa = 3.42RR239 pKa = 11.84NTRR242 pKa = 11.84YY243 pKa = 10.27GRR245 pKa = 11.84LIGWW249 pKa = 3.97
Molecular weight: 28.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5979 |
184 |
1219 |
498.3 |
56.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.138 ± 0.638 | 1.254 ± 0.199 |
6.673 ± 0.358 | 5.135 ± 0.211 |
3.897 ± 0.243 | 5.503 ± 0.253 |
2.342 ± 0.222 | 6.857 ± 0.286 |
6.573 ± 0.37 | 9.282 ± 0.389 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.726 ± 0.112 | 6.958 ± 0.371 |
3.278 ± 0.315 | 3.061 ± 0.38 |
4.7 ± 0.284 | 7.242 ± 0.456 |
6.004 ± 0.504 | 7.459 ± 0.295 |
0.669 ± 0.125 | 4.248 ± 0.222 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
